| Carnitine O-octanoyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 2.3.1.137 | ||||||||
| CAS no. | 39369-19-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Carnitine O-octanoyltransferase (CROT or COT) is a member of the transferase family, more specifically a carnitine acyltransferase, a type of enzyme which catalyzes the transfer of acyl groups from acyl-CoAs to carnitine, generating CoA and an acyl-carnitine. (EC 2.3.1.137) The systematic name of this enzyme is octanoyl-CoA:L-carnitine O-octanoyltransferase. Other names in common use include medium-chain/long-chain carnitine acyltransferase, carnitine medium-chain acyltransferase, easily solubilized mitochondrial carnitine palmitoyltransferase, and overt mitochondrial carnitine palmitoyltransferase. Specifically, CROT catalyzes the chemical reaction:
- octanoyl-CoA + L-carnitine CoA + L-octanoylcarnitine
Thus, the two substrates of this enzyme are octanoyl-CoA and L-carnitine and its two products are CoA and L-octanoylcarnitine.[1]
This reaction is easily chemically reversible, and does not require any energy input, as both fatty acyl-CoAs and fatty acylcarnitines are considered chemically “activated” forms of fatty acyl groups.[2]
This enzyme belongs to the family of transferases, specifically those acyltransferases transferring groups other than aminoacyl groups.
Structure

CROT is 612 amino acids long,[3] with a molecular weight of about 70 kDa.[2] In terms of broad overall structural features, CROT has 20 α-helices and 16 β-strands, and can be divided into two overall domains, named N and C.[3]
As of late 2007, 4 structures have been solved for this class of enzymes, with PDB accession codes 1XL7, 1XL8, 1XMC, and 1XMD.
The significant catalytic residue within all carnitine acyltransferases, including CROT, has been found to be a histidine residue, confirmed by site-directed mutagenesis studies. In CROT, this residue is at position 327. This residue, including the rest of the active site, is found at the interface between the N and C domains. The active site stabilizes carnitine by an intricate network of hydrogen-bonding residues, along with a key water molecule. The longer acyl chain is stabilized by hydrophobic residues arrayed in an approximately-cylindrical fashion.[3]
As may be expected from members of the same enzymatic family, there is strong similarity between the structures of carnitine acetyltransferase (CRAT) and CROT, as these enzymes have 36% sequence homology.[3] A key difference between these enzymes that may explain their selectivities between short and medium-chain acyl-CoAs hinges on a glycine residue which is present in the acyl binding site in CROT, Gly-553. In CRAT, however, the residue in the same position in the acyl binding site is a methionine residue, Met-564.[3] These residues has been shown to serve as a substrate “gatekeeper” in both CRAT and CROT. M564G CRAT mutants have been shown to accept a wider variety of acyl-CoA substrates.[4] Similarly, G553M CROT mutants show marked inactivity with octanoyl-CoA, while maintaining activity with short-chain acyl-CoAs.[3]
Function
One function of CROT is to supply acetyl-CoA to glucose-starved cells. In the absence of carnitine acetyltransferase (CRAT), acyltransferases such as CROT can catalyze the acetyl group transfer from acetylcarnitine to coenzyme A. Rescue experiments with CROT gene knockout cells have shown that peroxisomal CROT can mediate acetyl-CoA production under glucose-limited conditions. The peroxisome can then export these products into the cytosol.[5]
Localization
Though CROT is distributed on both sides of microsomal vesicles, it has also been found that the bulk of CROT activity in murine liver is in the cytoplasm face of the vesicles and endoplasmic reticulum. CROT may play a role in converting peroxisomal medium-chain acylcarnitine derivatives to medium-chain acyl-CoA derivatives. These can then feed into a variety of biosynthetic pathways for elongations and other modifications.[6]
In addition, CROT is inhibited by trypsin in a dose-dependent manner. A maximum of 60% inhibition was observed in purified CROT, similar to what was seen with carnitine palmitoyltransferase (CPT). CROT activity also appears to be inhibited to the same extent in both permeable and sealed microsomal membranes.[6]
CROT is thought to be peroxisomally-located. It was found that administration of di(2-ethylhexyl)phthalate (DEHP), a peroxisomal proliferator, to Wistar rats led to an increase in the expression of CROT by a factor of 14.1. This was as a result of increased translation of CROT mRNA, along with decreased degradation by a factor of 1.5.[7]
CROT activity has also been reported in mouse liver, kidney, adipocyte, mammary gland, skeletal muscle, and heart tissues. It was found that CROT activity in the kidney was mostly overt, while in the liver and heart it was mainly latent. Interestingly, the trend for a related enzyme, carnitine palmitoyl transferase (CPT), the opposite trend was found.[8]
Substrates
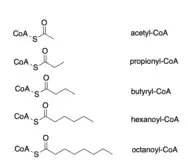
While CROT's canonical substrate is octanoyl-CoA, CROT is also known to be able to catalyze the deacylation of numerous acyl-CoAs, such as acetyl-CoA, propionyl-CoA, butyryl-CoA, and hexanoyl-CoA.[5] CROT can also take branched-chain fatty acyl-CoAs as substrates, such as 4,8-dimethylnonanoyl-CoA, which is derived from the metabolism of pristanic acid in the peroxisome.[9]
Regulation
Because CROT activity has a role in beta-oxidation of fatty acids and ketone body synthesis, it is an important point of regulation. One known inhibitor of CROT is malonyl-CoA, which inhibits CROT non-linearly. Complex kinetic behavior is observed when malonyl-CoA is incubated with purified CROT.
A decrease in pH can also enhance malonyl-CoA inhibition of CROT. Some studies have indicated that when the pH of assaying conditions was decreased from 7.4 to 6.8, inhibition could increase by 20-30%. Further, the Ki for malonyl-CoA in CROT decreases from 106 uM to 35 uM over this drop. This change is not seen for palmitoyl-CoA and decanoyl-CoA. However, the degree of inhibition by malonyl-CoA is similar to that observed with other short-chain acyl-CoA esters, such as glutaryl-CoA, hydroxymethylglutaryl-CoA, and methylmalonyl-CoA.
The ionization state of malonyl-CoA does not change significantly over the pH range 7.4-6.8. The change in sensitivity to inhibitors may be due to the CROT active-site His-327 residue. Malonyl-CoA is also found at a lower concentration in the cell (1-6 uM) than its Ki.Thus, its inhibition of CROT may not be physiologically significant under homeostatic conditions.[10]
References
- ↑ Farrell, S. O.; Fiol, C. J.; Reddy, J. K.; Bieber, L. L. (1984-11-10). "Properties of purified carnitine acyltransferases of mouse liver peroxisomes". The Journal of Biological Chemistry. 259 (21): 13089–13095. doi:10.1016/S0021-9258(18)90661-7. ISSN 0021-9258. PMID 6436243.
- 1 2 Jogl, Gerwald; Hsiao, Yu-Shan; Tong, Liang (November 2004). "Structure and function of carnitine acyltransferases". Annals of the New York Academy of Sciences. 1033 (1): 17–29. Bibcode:2004NYASA1033...17J. doi:10.1196/annals.1320.002. ISSN 0077-8923. PMID 15591000. S2CID 24466239.
- 1 2 3 4 5 6 Jogl, Gerwald; Hsiao, Yu-Shan; Tong, Liang (2005-01-07). "Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity". The Journal of Biological Chemistry. 280 (1): 738–744. doi:10.1074/jbc.M409894200. ISSN 0021-9258. PMID 15492013.
- ↑ Hsiao, Yu-Shan; Jogl, Gerwald; Tong, Liang (2004-07-23). "Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase". The Journal of Biological Chemistry. 279 (30): 31584–31589. doi:10.1074/jbc.M403484200. ISSN 0021-9258. PMID 15155726.
- 1 2 Hsu, Jake; Fatuzzo, Nina; Weng, Nielson; Michno, Wojciech; Dong, Wentao; Kienle, Maryline; Dai, Yuqin; Pasca, Anca; Abu-Remaileh, Monther; Rasgon, Natalie; Bigio, Benedetta; Nasca, Carla; Khosla, Chaitan (February 2023). "Carnitine octanoyltransferase is important for the assimilation of exogenous acetyl-L-carnitine into acetyl-CoA in mammalian cells". The Journal of Biological Chemistry. 299 (2): 102848. doi:10.1016/j.jbc.2022.102848. ISSN 1083-351X. PMC 9898754. PMID 36587768.
- 1 2 Valkner, K. J.; Bieber, L. L. (1982-07-14). "The sidedness of carnitine acetyltransferase and carnitine octanoyltransferase of rat liver endoplasmic reticulum". Biochimica et Biophysica Acta (BBA) - Biomembranes. 689 (1): 73–79. doi:10.1016/0005-2736(82)90190-0. ISSN 0006-3002. PMID 7104352.
- ↑ Ozasa, H.; Miyazawa, S.; Osumi, T. (August 1983). "Biosynthesis of carnitine octanoyltransferase and carnitine palmitoyltransferase". Journal of Biochemistry. 94 (2): 543–549. doi:10.1093/oxfordjournals.jbchem.a134385. ISSN 0021-924X. PMID 6630174.
- ↑ Saggerson, E. D.; Carpenter, C. A. (1981-07-06). "Carnitine palmitoyltransferase and carnitine octanoyltransferase activities in liver, kidney cortex, adipocyte, lactating mammary gland, skeletal muscle and heart". FEBS Letters. 129 (2): 229–232. doi:10.1016/0014-5793(81)80171-8. ISSN 0014-5793. PMID 7286216. S2CID 2011527.
- ↑ Ferdinandusse, S.; Mulders, J.; IJlst, L.; Denis, S.; Dacremont, G.; Waterham, H. R.; Wanders, R. J. (1999-09-16). "Molecular cloning and expression of human carnitine octanoyltransferase: evidence for its role in the peroxisomal beta-oxidation of branched-chain fatty acids". Biochemical and Biophysical Research Communications. 263 (1): 213–218. doi:10.1006/bbrc.1999.1340. ISSN 0006-291X. PMID 10486279.
- ↑ A'Bháird, N. N.; Ramsay, R. R. (1992-09-01). "Malonyl-CoA inhibition of peroxisomal carnitine octanoyltransferase". The Biochemical Journal. 286 ( Pt 2) (Pt 2): 637–640. doi:10.1042/bj2860637. ISSN 0264-6021. PMC 1132947. PMID 1530596.