| Thiosulfate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
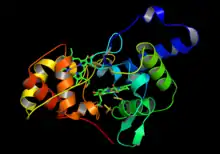 3D structure of thiosulfate dehydrogenase with thiosulfate substrate present in the active site, corresponding to RCSB code 4V2K | |||||||||
| Identifiers | |||||||||
| EC no. | 1.8.2.2 | ||||||||
| CAS no. | 9076-88-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Thiosulfate dehydrogenase (abbreviated as TsdA) (EC 1.8.2.2) is an enzyme that catalyzes the chemical reaction:[1]
- 2 thiosulfate + 2 ferricytochrome c tetrathionate + 2 ferrocytochrome c
Thus, the two substrates of this enzyme are thiosulfate and ferricytochrome c, whereas its two products are tetrathionate and ferrocytochrome c.
Thiosulfate dehydrogenase homologues have been isolated from numerous bacterial species and differ slightly in structure but have analogous function and mechanism of sulfur oxidation. The enzyme is similar in both function and structure to a few enzymes in the Sox sulfur oxidation pathway.[2]
Nomenclature
This enzyme belongs to the family of oxidoreductases, specifically those acting on a sulfur group of donors with a cytochrome as acceptor. The systematic name of this enzyme class is thiosulfate:ferricytochrome-c oxidoreductase. Other names in common use include tetrathionate synthase, thiosulfate oxidase, thiosulfate-oxidizing enzyme, and thiosulfate-acceptor oxidoreductase.
Structure
Thiosulfate dehydrogenase, isolated from the appreciably studied bacterial strain Allochromatium vinosum (253 peptide chain length, 25.8 kDa) is composed of two catalytic domains, each similar to cytochrome c, linked by a long unstructured peptide chain.[3] The N-terminal domain is structurally homologous to the SoxA family of cytochrome enzymes while the C-terminal domain is representative of the standard mitochondrial cytochrome c family fold with high similarity to nitrite reductase from P. haloplanktis.[4] Each domain contains a covalently bound iron-containing heme molecule separated by a short distance of 8.1 Å which assists with rapid electron transfer.[5] Both the N and C terminus domains contain 4 α helices (surrounding the heme in the corresponding domain) and a two-stranded anti-parallel β sheet, suggesting the enzyme resulted from a gene duplication event.[6]
The single active site of the enzyme is located in between the two domains (closer to the C-terminus domain) near the central iron heme.
Mechanism
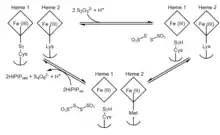
There is controversy to the exact mechanism that the enzyme enables to occur, so the process remains ambiguous. Additionally, the variety of thiosulfate dehydrogenase enzymes among bacterial species implies several possible mechanisms of activity.[5] However, due to the striking similarity in structure the domains of thiosulfate dehydrogenase have to sulfur carrier protein SoxYZ and cytochrome SoxAX, a related mechanism can be derived for the thiosulfate dehydrogenase-catalyzed reaction in A. vinosum. The overall, generalized overview of the proposed mechanism of thiosulfate dehydrogenase can be summarized by the following two reversible redox reactions:
Step 1 indicates an initial binding of thiosulfate to an unusual and reactive cysteine S-sulphane adduct, forming a S-thiosulfonate adduct. Step 2 follows with an additional thiosulfate to subsequently form tetrathionate while reducing both hemes and leaving a typical cysteine residue. In the active site, the cysteine residue bound to the catalytic iron heme is essential for enzymatic activity, as the abolishment of this residue completely eliminated the enzyme's ability to both oxidize thiosulfate and reduce tetrathionate.[7] In A. vinosum, although the process is reversible, the reaction to form two thiosulfate ions is much slower than the formation of tetrathionate despite the reduction of tetrathionate to thiosulfate having a potential of +198 mV.[8]
Reduction of the enzyme results in a ligand switch from Lys208 to Met209 in the second heme. Mutant proteins that replace Met209 with asparagine or glycine have similar substrate affinities to the wildtype variant but have much lower specific activities, suggesting that heme 2 is the electron exit point in the last steps of the mechanism.[9] Upon the reduction of heme 2 and the ligand switch, the redox potential is increased and hinders the back reaction to form thiosulfate. Here, it is suggested that a high potential iron-sulfur protein (HiPIP) serves as the electron acceptor in the oxidation of both hemes to their initial state.
Function
The oxidation of thiosulfate to tetrathionate is observed in several thiobacilli, phototrophs, and heterotrophs, as thiosulfate and tetrathionate play the role of electron donor and electron acceptor, respectively, in many bacterial species.[10] Both compounds are intermediates in and play an important role in the biogeochemical sulfur cycle, the process of conversion between sulfide and sulfate.[11] Thus, thiosulfate dehydrogenase is essential for the conversion between the intermediates in the sulfur cycle.[12] The sulfur cycle enables a variety of bacteria to utilize generated thiosulfate as an electron donor for aerobic growth and anaerobic carbon dioxide fixation for photosynthesis. Pseudomonas and Halomonas are examples of the many thiobacteria that utilize thiosulfate dehydrogenase to derive energy from thiosulfate as a supplemental energy source.[13] Tetrathionate can serve as a respiratory electron acceptor during anaerobic respiration by tetrathionate reduction.
Industrial applications
Thiobacteria such as Acidithiobacillus ferrooxidans have become essential to industrial bioleaching applications, as the microorganisms are able to oxidize iron and sulfur from iron-sulfur minerals as energy sources, supporting their own autotrophic growth while producing ferric iron and sulfuric acid.[14] Thus, bacteria have been isolated from mineral deposits and used in the treatment of refractory gold and iron ores and detoxification of industrial waste products, sewage, and soils contaminated with heavy metals.[15]
References
- ↑ Lu WP, Kelly DP (1988). "Cellular Location and Partial Purification of the 'Thiosulphate-oxidizing Enzyme' and 'Trithionate Hydrolyase' from Thiobacillus tepidarius". Microbiology. 134 (4): 877–885. doi:10.1099/00221287-134-4-877.
- ↑ Bamford, V. A. (2002). "Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme". The EMBO Journal. 21 (21): 5599–5610. doi:10.1093/emboj/cdf566. ISSN 1460-2075. PMC 131063. PMID 12411478.
- ↑ PDB: 4V2K; Grabarczyk DB, Chappell PE, Eisel B, Johnson S, Lea SM, Berks BC (April 2015). "Mechanism of thiosulfate oxidation in the SoxA family of cysteine-ligated cytochromes". The Journal of Biological Chemistry. 290 (14): 9209–21. doi:10.1074/jbc.M114.618025. PMC 4423706. PMID 25673696.
- ↑ Grabarczyk DB, Chappell PE, Eisel B, Johnson S, Lea SM, Berks BC (April 2015). "Mechanism of thiosulfate oxidation in the SoxA family of cysteine-ligated cytochromes". The Journal of Biological Chemistry. 290 (14): 9209–21. doi:10.1074/jbc.M114.618025. PMC 4423706. PMID 25673696.
- 1 2 Kurth JM, Brito JA, Reuter J, Flegler A, Koch T, Franke T, Klein EM, Rowe SF, Butt JN, Denkmann K, Pereira IA, Archer M, Dahl C (November 2016). "Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase". The Journal of Biological Chemistry. 291 (48): 24804–24818. doi:10.1074/jbc.M116.753863. PMC 5122753. PMID 27694441.
- ↑ Brito JA, Denkmann K, Pereira IA, Archer M, Dahl C (April 2015). "Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation". The Journal of Biological Chemistry. 290 (14): 9222–38. doi:10.1074/jbc.M114.623397. PMC 4423707. PMID 25673691.
- ↑ Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IA, Dahl C (October 2012). "Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome". Environmental Microbiology. 14 (10): 2673–88. doi:10.1111/j.1462-2920.2012.02820.x. PMID 22779704.
- ↑ Kurth JM, Dahl C, Butt JN (October 2015). "Catalytic Protein Film Electrochemistry Provides a Direct Measure of the Tetrathionate/Thiosulfate Reduction Potential". Journal of the American Chemical Society. 137 (41): 13232–5. doi:10.1021/jacs.5b08291. PMID 26437022.
- ↑ Brito JA, Gutierres A, Denkmann K, Dahl C, Archer M (October 2014). "Production, crystallization and preliminary crystallographic analysis of Allochromatium vinosum thiosulfate dehydrogenase TsdA, an unusual acidophilic c-type cytochrome". Acta Crystallographica Section F. 70 (Pt 10): 1424–7. doi:10.1107/S2053230X14019384. PMC 4188095. PMID 25286955.
- ↑ Kelly DP, Wood AP (1994). "Chapter 36: Enzymes involved in microbiological oxidation of thiosulfate and polythionates". In Peck HD, LeGall J (eds.). Inorganic Microbial Sulfur Metabolism. Methods in Enzymology. Vol. 243. pp. 501–510. doi:10.1016/0076-6879(94)43038-1. ISBN 978-0-12-182144-9.
- ↑ Zopfi J, Ferdelman TG, Fossing H (2004). "Distribution and fate of sulfur intermediates—sulfite, tetrathionate, thiosulfate, and elemental sulfur—in marine sediments". In Amend JP, Edwards KJ, Lyons TW (eds.). Sulfur Biogeochemistry - Past and Present. Vol. 379. Geological Society of America. pp. 97–116. doi:10.1130/0-8137-2379-5.97. ISBN 978-0-8137-2379-2.
- ↑ Sievert S, Kiene R, Schulz-Vogt H (2007). "The Sulfur Cycle". Oceanography. 20 (2): 117–123. doi:10.5670/oceanog.2007.55. hdl:1912/2786.
- ↑ Podgorsek L, Imhoff JF (1999). "Tetrathionate production by sulfur oxidizing bacteria and the role of tetrathionate in the sulfur cycle of Baltic Sea sediments" (PDF). Aquatic Microbial Ecology. 17: 255–265. doi:10.3354/ame017255.
- ↑ Bosecker K (1997). "Bioleaching: metal solubilization by microorganisms". FEMS Microbiology Reviews. 20 (3–4): 591–604. doi:10.1111/j.1574-6976.1997.tb00340.x. S2CID 86689062.
- ↑ Rawlings DE (May 2005). "Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates". Microbial Cell Factories. 4 (1): 13. doi:10.1186/1475-2859-4-13. PMC 1142338. PMID 15877814.