| Inositol trisphosphate 3-kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Inositol-trisphosphate 3-kinase A Catalytic Core. 1TZD | |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.1.127 | ||||||||
| CAS no. | 106283-10-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Inositol (1,4,5) trisphosphate 3-kinase (EC 2.7.1.127), abbreviated here as ITP3K, is an enzyme that facilitates a phospho-group transfer from adenosine triphosphate to 1D-myo-inositol 1,4,5-trisphosphate. This enzyme belongs to the family of transferases, specifically those transferring phosphorus-containing groups (phosphotransferases) with an alcohol group as acceptor. The systematic name of this enzyme class is ATP:1D-myo-inositol-1,4,5-trisphosphate 3-phosphotransferase. ITP3K catalyzes the transfer of the gamma-phosphate from ATP to the 3-position of inositol 1,4,5-trisphosphate to form inositol 1,3,4,5-tetrakisphosphate.[1] ITP3K is highly specific for the 1,4,5-isomer of IP3, and it exclusively phosphorylates the 3-OH position, producing Ins(1,3,4,5)P4, also known as inositol tetrakisphosphate or IP4.
In biology, the enzyme ITP3K is abbreviated a number of different ways, including 1D-myo-inositol-trisphosphate 3-kinase, ITP3K, ITPK, IP3-kinase, IP3-3-kinase, Ins(1,4,5)P3 3-kinase. In addition the enzyme may be named as the product of one of 3 genes in humans ITPKA, ITPKB, and ITPKC, or one of two in fruit flies, IP3K1 and IP3K2—a mutant known to geneticists as wavy.[2] The nematode genome has one form of the enzyme, coded by the LFE-2 gene. ITP3K enzymes are expressed only in metazoans; they are not expressed in yeast or plants.
All ITP3Ks belong to a larger structural family, the inositol polyphosphate kinases, or IPKs. Note however, that the human genome also contains a gene for a different kinase known as ITPK1, which is an inositol 1, 3, 4-trisphosphate 5/6-kinase and is not a member of the IPK family.
The ITP3K enzyme family is sometimes confused with a different enzyme family that has a similar name, that is, the phosphatidyl inositol 3-kinases or phosphoinositide 3-kinase (PI3-K),whose substrates are inositol lipids, not the soluble second messenger inositol trisphosphate.
Discovery and characterization
Scientific interest in the inositol phosphates intensified in the years following the 1983 discovery that inositol trisphosphate was an intracellular messenger that releases calcium from intracellular stores in the endoplasmic reticulum.[3] By the end of the decade, a large number of inositol phosphate kinases and phosphatases had been discovered, including ITP3K in 1986.[4][5] Biochemical and molecular studies in the 1990s led to the purification of the enzyme from rat brain and it molecular cloning, and these studies revealed various feedback mechanisms by which the enzyme is regulated by calcium and protein kinases.[6] In 1999, ITP3K was identified as being a member of a larger family of Inositol polyphosphate kinases, which share a similar structure and catalytic mechanism.[7][8] ITP3K enzymes share common structural features including a conserved catalytic core which binds ATP located near the C-terminus, and various regulatory domains nearer to the N-terminus.[9]
Catalytic domain
Evidence for this exquisite specificity and for the catalytic mechanism was found when the apo-enzyme, substrate-bound complex, and product-bound complex X-ray crystal structures of ITPKA were determined.[10][11] The figure to the right depicts the catalytic mechanism, whereby the 3'OH of IP3 attacks the gamma-phosphate of ATP, and amino acid residues of ITPK important for stabilizing the substrates and products in the active site.
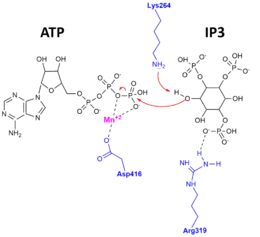
The structure of the catalytic domain of the human ITP3KA has been shown to be divided into three subdomains. These subdomains are displayed as the N lobe, which is a N-terminal domain, the C lobe, which is a C-terminal subdomain and a third alpha-only subdomain. The ITP3K catalytic domain varies somewhat from the protein kinase superfamily, and it has a novel four-helix substrate binding domain. In this kinase, the two domains are in an open conformation, which indicates that the two domains are both accessible at the same time. This suggests that substrate recognition and catalysis by ITP3K involves a dynamic conformational cycle. Additionally, this unique helical domain of ITPK blocks access to the active site by membrane-bound phosphoinositides, explaining the structural basis for soluble inositol polyphosphate specificity. Another feature of the catalytic core is the ATP binding site. Here, one molecule of ADP is bound in the cleft of the major domain, which indicates the active site of the kinase.
In further detail, the larger domain of the protein structure has an α/β-class structure. The domain has an N-terminal and a C-terminal lobe with a cleft in between and each of these lobes is built around an antiparallel β-sheet. In the N-terminal, the sheet has three strands, whereas in the C-terminal there is a five-stranded sheet. The second domain, is α-helical and consists of four α helices linked by long loops. The helices are loosely packed against each other and the entire domain is highly mobile as compared to the large α/β domain. The helical domain is juxtaposed against one end of the cleft in the large domain.
Regulation
ITP3K is regulated by various post-translational mechanisms. ITP3Ks are stimulated directly by calcium/calmodulin (Ca2+/CaM) binding.[12] Generally, mammalian ITP3Ks are activated by calcium and calmodulin to varying degrees. The method in which this works is calmodulin recognizes sequences which contain amphiphilic alpha-helices with clusters of positively charged and hydrophobic amino acids.[13] Certain sequences are required for CaM binding and enzyme activation and this level of stimulation appears to be specific to cell, tissue, or isoform. ITP3Ks from nematodes and Arabidopsis thaliana lack the CaM-binding sites and therefore are insensitive to calcium and calmodulin.[14] Another major post-translational modification that is important for ITP3K regulation is phosphorylation. ITP3K activity is indirectly stimulated by phosphorylation by calcium/calmodulin-dependent kinase II (CaMKII). In addition, there is evidence that ITP3Ks may be activated upon phosphorylation by protein kinase C (PKC) and inhibited upon phosphorylation by protein kinase A (PKA).
Isoforms
There are three ITP3Ks which are encoded by the human genome: ITPKA, ITPKB, and ITPKC. All share a conserved C-terminal catalytic domain, but differ in mechanisms of regulation as well as tissue expression. ITPKA is predominant in neurons and in the testes. It is localized to dendritic spines by an association with filamentous actin which is consistent with its probable role in memory functions. ITPKB is expressed more widely, but it is often enriched in immune tissues, and it has different intracellular localizations that depend on tissue, interaction with actin filaments, and proteolysis at the N-terminal regions. ITPKC is also expressed in many different tissues and it is more enriched in the nucleus compared to the other isoforms.
Functions in Calcium Signaling
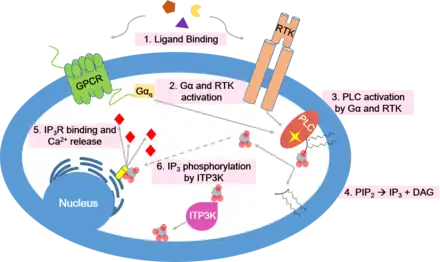
ITP3K plays a role in regulating or cooperating with intracellular calcium signals that occur following the liberation of inositol trisphosphate. In this pathway, either a G-protein coupled receptor (GPCR) or receptor tyrosine kinase (RTK) is activated by an extracellular ligand-binding event. Initiation of the pathway leads to an activated G-alpha subunit of a heterotrimeric G protein (in the case of GPCR-mediated signal transduction) or autophoshorylation of RTK cytoplasmic domains (in the case of RTK-mediated signal transduction). These intracellular events eventually lead to activation of phospholipase C (PLC), which cleaves the phospholipid PIP2 into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). DAG remains associated with the plasma membrane, while IP3 is released into the cytoplasm. IP3 then diffuses through the cytosol and binds to IP3 receptors on the endoplasmic reticulum or sarcoplasmic reticulum, resulting in the opening of a membrane channel and an influx of calcium ions into the cytoplasm.[15] Calcium serves as a second messenger for various downstream cellular events including glycogen metabolism, muscle contraction, neurotransmitter release, and transcriptional regulation.[15] Therefore, calcium homeostasis is essential for proper cell function and response to extracellular signals.[16]
In order to prepare the cell for a future signaling event, the calcium pathway must be tightly regulated. ITP3K seems to play an important role in termination of the signal. As mentioned, ITP3K catalyzes the phosphorylation of IP3 to make IP4. Unlike IP3, IP4 does not cause opening of calcium channels on the endoplasmic reticulum or sarcoplasmic reticulum.[17] By decreasing the concentration of IP3 in the cytoplasm, ITP3K terminates propagation of the calcium signaling pathway.[14]
Additional roles
ITP3K is not the only enzyme responsible for clearing IP3 from the cytoplasm. A second enzyme called inositol 5-phosphatase catalyzes the dephosphorylation of IP3 to create IP2.[18] Typically, nature does not favor the evolution of a second enzyme to perform an already-existing, identical function.[19] A closer inspection of the evolutionary history of inositol 5-phosphatase and ITP3K gives rise to several interesting hypotheses about the roles of these enzymes in the cell.
Inositol 5-phosphatase existed before ITP3K evolved in the mammalian cell. Like other phosphatases, inositol 5-phosphatase is an energy-independent enzyme that cleaves a phosphate group off of a substrate.[20] In contrast, ITP3K (like all kinases) is energy-dependent, meaning that it requires an ATP molecule to perform the phosphoryl transfer chemistry.[21] If nature already had an energy-independent mechanism for termination of the calcium signaling pathway, why was the evolution of ITP3K advantageous? This apparent redundancy of function, or "waste" of energy by the cell, suggests that ITP3K may have a more important function in the cell than simply clearing the IP3 second messenger from the cytoplasm.[20] Current hypotheses about additional roles for ITPK are explained in the following two subsections.
Product of ITPK may be a second messenger
As mentioned previously, ITP3K catalyzes a phosphoryl transfer reaction that converts IP3 to IP4. IP4 does not stimulate calcium influx through IP3 receptor channels on the endoplasmic or sarcoplasmic reticulum. However, it has been shown that IP4 stimulates calcium channel opening on the plasma membrane. In this way, IP4 may actually serve to prolong the calcium signal by activating the influx of calcium stores from the extracellular space. In addition, there is evidence that IP4 binds two GTPase-activating proteins, GAP1IP4BP and GAP1m.[18] GAPs are often used in signal transduction as on/off switches. IP4 binding to GAPs suggests that ITPK may be involved in a parallel signal transduction pathway. The exact role of IP4 binding to these GAPs has not been determined, though, so additional research in this area will be needed to gain a more complete understanding.[22]
Role in inositol phosphate metabolism
In addition to its potential roles as a second messenger, IP4 may also function as an essential precursor for other more highly phosphorylated inositol phosphates such as IP5, IP6, IP7, and IP8. Such maintenance is necessary to prepare the cell for a future incoming signal.[22]
Relevance to physiology and human disease
ITPKA protein is highly enriched in dendritic spines.[23] ITPKA participates in learning and memory process in neuronal cells, both via its catalytic activity and its interaction with filamentous actin.
Although ITPKA is expressed physiologically in neurons and testis, the gene becomes expressed in a number of cancer cell types. In most cases, ITP3K expression causes the cancer to be more aggressive.[24]
ITPKB is implicated in physiologic immune function.[25]
ITPKC has been linked to Kawasaki Disease, an autoimmune disorder.[26][27]
References
- ↑ "UniProtKB - P23677 (IP3KA_HUMAN)". Retrieved 19 February 2015.
- ↑ Dean DM, Maroja LS, Cottrill S, Bomkamp BE, Westervelt KA, Deitcher DL (November 2015). "The wavy Mutation Maps to the Inositol 1,4,5-Trisphosphate 3-Kinase 2 (IP3K2) Gene of Drosophila and Interacts with IP3R to Affect Wing Development". G3. 6 (2): 299–310. doi:10.1534/g3.115.024307. PMC 4751550. PMID 26613949.
- ↑ Streb H, Irvine RF, Berridge MJ, Schulz I (1983). "Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate". Nature. 306 (5938): 67–9. Bibcode:1983Natur.306...67S. doi:10.1038/306067a0. PMID 6605482. S2CID 4359904.
- ↑ Irvine RF, Letcher AJ, Heslop JP, Berridge MJ (1986). "The inositol tris/tetrakisphosphate pathway--demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues". Nature. 320 (6063): 631–4. Bibcode:1986Natur.320..631I. doi:10.1038/320631a0. PMID 3010126. S2CID 4249596.
- ↑ Hansen CA, Mah S, Williamson JR (June 1986). "Formation and metabolism of inositol 1,3,4,5-tetrakisphosphate in liver". The Journal of Biological Chemistry. 261 (18): 8100–3. doi:10.1016/S0021-9258(19)83881-4. PMID 3487541.
- ↑ Takazawa K, Vandekerckhove J, Dumont JE, Erneux C (November 1990). "Cloning and expression in Escherichia coli of a rat brain cDNA encoding a Ca2+/calmodulin-sensitive inositol 1,4,5-trisphosphate 3-kinase". The Biochemical Journal. 272 (1): 107–12. doi:10.1042/bj2720107. PMC 1149663. PMID 2176078.
- ↑ Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH (November 1999). "Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases". Current Biology. 9 (22): 1323–6. doi:10.1016/s0960-9822(00)80055-x. PMID 10574768. S2CID 15311443.
- ↑ Odom AR, Stahlberg A, Wente SR, York JD (March 2000). "A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control". Science. 287 (5460): 2026–9. Bibcode:2000Sci...287.2026O. doi:10.1126/science.287.5460.2026. PMID 10720331.
- ↑ Schell MJ (June 2010). "Inositol trisphosphate 3-kinases: focus on immune and neuronal signaling". Cellular and Molecular Life Sciences. 67 (11): 1755–78. doi:10.1007/s00018-009-0238-5. PMID 20066467. S2CID 25121695.
- ↑ González B, Schell MJ, Letcher AJ, Veprintsev DB, Irvine RF, Williams RL (September 2004). "Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase". Molecular Cell. 15 (5): 689–701. doi:10.1016/j.molcel.2004.08.004. PMID 15350214.
- ↑ Miller GJ, Hurley JH (September 2004). "Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase". Molecular Cell. 15 (5): 703–11. doi:10.1016/j.molcel.2004.08.005. PMID 15350215.
- ↑ Lloyd-Burton SM, Yu JC, Irvine RF, Schell MJ (March 2007). "Regulation of inositol 1,4,5-trisphosphate 3-kinases by calcium and localization in cells". The Journal of Biological Chemistry. 282 (13): 9526–35. doi:10.1074/jbc.M610253200. PMID 17284449.
- ↑ Franco-Echevarría E, Baños-Sanz JI, Monterroso B, Round A, Sanz-Aparicio J, González B (November 2014). "A new calmodulin-binding motif for inositol 1,4,5-trisphosphate 3-kinase regulation". The Biochemical Journal. 463 (3): 319–28. doi:10.1042/BJ20140757. PMID 25101901.
- 1 2 Xia HJ, Yang G (February 2005). "Inositol 1,4,5-trisphosphate 3-kinases: functions and regulations". Cell Research. 15 (2): 83–91. doi:10.1038/sj.cr.7290270. PMID 15740635.
- 1 2 Berridge MJ (January 1993). "Inositol trisphosphate and calcium signalling". Nature. 361 (6410): 315–25. Bibcode:1993Natur.361..315B. doi:10.1038/361315a0. PMID 8381210. S2CID 4362262.
- ↑ Voet, Donald Voet, Judith G. (2011). Biochemistry (4th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 978-0-470-57095-1.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ↑ Havas N (August 2011). "Back in the water". Journal of Palliative Medicine. 14 (8): 971–2. doi:10.1089/jpm.2011.0043. PMID 21809925.
- 1 2 Pattni K, Banting G (June 2004). "Ins(1,4,5)P3 metabolism and the family of IP3-3Kinases". Cellular Signalling. 16 (6): 643–54. doi:10.1016/j.cellsig.2003.10.009. PMID 15093605.
- ↑ "Understanding Evolution". Retrieved 19 February 2015.
- 1 2 Irvine RF, Lloyd-Burton SM, Yu JC, Letcher AJ, Schell MJ (2006). "The regulation and function of inositol 1,4,5-trisphosphate 3-kinases". Advances in Enzyme Regulation. 46 (1): 314–23. doi:10.1016/j.advenzreg.2006.01.009. PMC 1820747. PMID 16857241.
- ↑ "WikiKinome". Kinase.com. Retrieved 19 February 2015.
- 1 2 Irvine RF, Schell MJ (May 2001). "Back in the water: the return of the inositol phosphates". Nature Reviews. Molecular Cell Biology. 2 (5): 327–38. doi:10.1038/35073015. PMID 11331907. S2CID 2259401.
- ↑ Yamada M, Kakita A, Mizuguchi M, Rhee SG, Kim SU, Ikuta F (March 1993). "Specific expression of inositol 1,4,5-trisphosphate 3-kinase in dendritic spines". Brain Research. 606 (2): 335–40. doi:10.1016/0006-8993(93)91004-c. PMID 8387863. S2CID 10790958.
- ↑ Windhorst S, Fliegert R, Blechner C, Möllmann K, Hosseini Z, Günther T, Eiben M, Chang L, Lin HY, Fanick W, Schumacher U, Brandt B, Mayr GW (February 2010). "Inositol 1,4,5-trisphosphate 3-kinase-A is a new cell motility-promoting protein that increases the metastatic potential of tumor cells by two functional activities". The Journal of Biological Chemistry. 285 (8): 5541–54. doi:10.1074/jbc.M109.047050. PMC 2820782. PMID 20022963.
- ↑ Miller AT, Dahlberg C, Sandberg ML, Wen BG, Beisner DR, Hoerter JA, Parker A, Schmedt C, Stinson M, Avis J, Cienfuegos C, McPate M, Tranter P, Gosling M, Groot-Kormelink PJ, Dawson J, Pan S, Tian SS, Seidel HM, Cooke MP (2015). "Inhibition of the Inositol Kinase Itpkb Augments Calcium Signaling in Lymphocytes and Reveals a Novel Strategy to Treat Autoimmune Disease". PLOS ONE. 10 (6): e0131071. Bibcode:2015PLoSO..1031071M. doi:10.1371/journal.pone.0131071. PMC 4488288. PMID 26121493.
- ↑ Onouchi Y, Gunji T, Burns JC, Shimizu C, Newburger JW, Yashiro M, Nakamura Y, Yanagawa H, Wakui K, Fukushima Y, Kishi F, Hamamoto K, Terai M, Sato Y, Ouchi K, Saji T, Nariai A, Kaburagi Y, Yoshikawa T, Suzuki K, Tanaka T, Nagai T, Cho H, Fujino A, Sekine A, Nakamichi R, Tsunoda T, Kawasaki T, Nakamura Y, Hata A (January 2008). "ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms". Nature Genetics. 40 (1): 35–42. doi:10.1038/ng.2007.59. PMC 2876982. PMID 18084290.
- ↑ Alphonse MP, Duong TT, Shumitzu C, Hoang TL, McCrindle BW, Franco A, Schurmans S, Philpott DJ, Hibberd ML, Burns J, Kuijpers TW, Yeung RS (November 2016). "Inositol-Triphosphate 3-Kinase C Mediates Inflammasome Activation and Treatment Response in Kawasaki Disease". Journal of Immunology. 197 (9): 3481–3489. doi:10.4049/jimmunol.1600388. PMID 27694492.