| HindIII restriction endonuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | RE_Hindiii | ||||||||
| Pfam | PF09518 | ||||||||
| InterPro | IPR019043 | ||||||||
| |||||||||
| hindIIIR type II restriction endonuclease | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | hindIIIR | ||||||
| NCBI gene | 950303 | ||||||
| PDB | 2e52 More structures | ||||||
| UniProt | P43870 | ||||||
| Other data | |||||||
| EC number | 3.1.21.4 | ||||||
| |||||||
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.[1]

The cleavage of this sequence between the AA's results in 5' overhangs on the DNA called sticky ends:
5'-A |A G C T T-3'
3'-T T C G A| A-5'
Restriction endonucleases are used as defense mechanisms in prokaryotic organisms in the restriction modification system. Their primary function is to protect the host genome against invasion by foreign DNA, primarily bacteriophage DNA. There is also evidence that suggests the restriction enzymes may act alongside modification enzymes as selfish elements, or may be involved in genetic recombination and transposition.[2]
Enzyme Structure
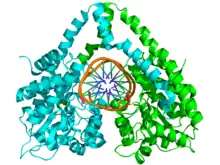
The structure of HindIII is complex, and consists of a homodimer. Like other type II restriction endonucleases, it is believed to contain a common structural core comprising four β-sheets and a single α-helix. Each subunit contains 300 amino acids and the predicted molecular mass is 34,950 Da. Despite the importance of this enzyme in molecular biology and DNA technology, little information is available concerning the mechanism of DNA recognition and phosphodiester bond cleavage.[1] However, it is believed that HindIII utilizes a common mechanism of recognition and catalysis of DNA found in other type II enzymes such as EcoRI, BamHI, and BglII. These enzymes contain the amino acid sequence motif PD-(D/E)XK to coordinate Mg2+, a cation required to cleave DNA in most type II restriction endonucleases.[4] The cofactor Mg2+ is believed to bind water molecules and carry them to the catalytic sites of the enzymes, among other cations. Unlike most documented type II restriction endonucleases, HindIII is unique in that it has little to no catalytic activity when Mg2+ is substituted for other cofactors, such as Mn2+.[1]
Site-directed mutagenesis
Despite the uncertainty concerning the structure-catalysis relationship of type II endonucleases, site-directed mutagenesis of the restriction endonuclease HindIII has provided much insight into the key amino acid residues involved. In particular, substitutions of Asn for Lys at residue 125 and Leu for Asp at residue 108 significantly decreased DNA binding and the catalytic function of HindIII.[1] In a separate mutagenesis study it was shown that a mutation at residue 123 from Asp to Asn reduced enzymatic activity. Despite the fact that this residue is most likely responsible for the unwinding of DNA and coordination to water rather than direct interaction with the attacking nucleophile, its specific function is unknown.[4]
Proposed mechanism
While restriction enzymes cleave at specific DNA sequences, they are first required to bind non-specifically with the DNA backbone before localizing to the restriction site. On average, the restriction enzyme will form 15-20 hydrogen bonds with the bases of the recognition sequence. With the aid of other van der Waals interactions, this bonding facilitates a conformational change of the DNA-enzyme complex which leads to the activation of catalytic centers.[2]
Despite the lack of evidence suggesting an exact mechanism for the cleavage of DNA by HindIII, site-mutagenesis analysis coupled with more detailed studies of metal ion-mediated catalysis in EcoRV have led to the following proposed catalytic mechanism. It has been suggested that during the hydrolysis of DNA by EcoRV the catalytic residue Lys-92 stabilizes and orients the attacking water nucleophile, while the carboxylate of Asp-90 stabilizes the leaving hydroxide anion through to coordination of Mg2+. Furthermore, enzymatic function is dependent upon the correct position of the Asp-74 residue, suggesting has a role in increasing the nucleophilicity of the attacking water molecule.[5]
As a result of the site-mutagenesis experiments previously outlined, it is thus proposed that Lys-125, Asp-123, and Asp-108 of HindIII function similarly to Lys-92, Asp-90, and Asp-74 in EcoRV, respectively. Lys-125 positions the attacking water molecule while Asp-108 improves its nucleophilicity. Asp-123 coordinates to Mg2+ which in turn stabilizes the leaving hydroxide ion.
Uses in research
HindIII as well as other type II restriction endonucleases are very useful in modern science, particularly in DNA sequencing and mapping. Unlike type I restriction enzymes, type II restriction endonucleases perform very specific cleaving of DNA. Type I restriction enzymes recognize specific sequences, but cleave DNA randomly at sites other than their recognition site whereas type II restriction enzymes cleave only at their specific recognition site.[6] Since their discovery in the early 1970s, type II restriction enzymes have revolutionized the way scientists work with DNA, particularly in genetic engineering and molecular biology.
Major uses of type II restriction enzymes include gene analysis and cloning. They have proven to be ideal modeling systems for the study of protein-nucleic acid interactions, structure-function relationships, and the mechanism of evolution.[2] They make good assays for the study of genetic mutations by their ability to specifically cleave DNA to allow the removal or insertion of DNA. Through the use of restriction enzymes, scientists are able to modify, insert, or remove specific genes, a very powerful tool especially when it comes to modifying an organism's genome.
References
- 1 2 3 4 Tang, D; et al. (2000). "Mutational analyses of restriction endonuclease-HindIII mutant E86K with higher activity and altered specificity". Protein Engineering. 13 (4): 283–9. doi:10.1093/protein/13.4.283. PMID 10810160.
- 1 2 3 Pingoud, Alfred; Jeltsch, Albert. (2001). "Structure and function of type II restriction endonucleases". Nucleic Acids Research. 29 (18): 3705–27. doi:10.1093/nar/29.18.3705. PMC 55916. PMID 11557805.
- ↑ Lukacs C, et al. (2000). "Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution". Nat. Struct. Biol. 7 (2): 134–40. doi:10.1038/72405. PMID 10655616. S2CID 20478739.
- 1 2 Tang D, et al. (1999). "Site-directed mutagenesis of restriction endonuclease HindIII". Biosci. Biotechnol. Biochem. 63 (10): 1703–7. doi:10.1271/bbb.63.1703. PMID 10586498.
- ↑ Horton N, Newberry K, Perona J (1999). "Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases". Proc. Natl. Acad. Sci. USA. 95 (23): 13489–94. doi:10.1073/pnas.95.23.13489. PMC 24846. PMID 9811827.
- ↑ Roberts, Richard J. (2005). "How restriction enzymes became the workhorses of molecular biology". Proc. Natl. Acad. Sci. USA. 102 (17): 5905–8. Bibcode:2005PNAS..102.5905R. doi:10.1073/pnas.0500923102. PMC 1087929. PMID 15840723.