| Epithelial sodium channel | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 Structure of human ENaC.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | ASC | ||||||||||
| Pfam | PF00858 | ||||||||||
| InterPro | IPR001873 | ||||||||||
| PROSITE | PDOC00926 | ||||||||||
| SCOP2 | 6BQN / SCOPe / SUPFAM | ||||||||||
| TCDB | 1.A.6 | ||||||||||
| OPM superfamily | 181 | ||||||||||
| OPM protein | 4fz1 | ||||||||||
| |||||||||||
The epithelial sodium channel (ENaC), (also known as amiloride-sensitive sodium channel) is a membrane-bound ion channel that is selectively permeable to sodium ions (Na+). It is assembled as a heterotrimer composed of three homologous subunits α or δ, β, and γ,[2] These subunits are encoded by four genes: SCNN1A, SCNN1B, SCNN1G, and SCNN1D. The ENaC is involved primarily in the reabsorption of sodium ions at the collecting ducts of the kidney's nephrons. In addition to being implicated in diseases where fluid balance across epithelial membranes is perturbed, including pulmonary edema, cystic fibrosis, COPD and COVID-19, proteolyzed forms of ENaC function as the human salt taste receptor.[3]
The apical membranes of many tight epithelia contain sodium channels that are characterized primarily by their high affinity for the diuretic blocker amiloride.[2][4][5][6] These channels mediate the first step of active sodium reabsorption essential for the maintenance of body salt and water homeostasis.[4] In vertebrates, the channels control reabsorption of sodium in kidney, colon, lung and sweat glands; they also play a role in taste perception.
The epithelial sodium channels are structurally and probably evolutionary related to P2X purinoreceptors, pain receptors that activate when they detect ATP.
Location and function
ENaC is located in the apical membrane of polarized epithelial cells in particular in the kidney (primarily in the collecting tubule), the lung, the skin,[7] the male and female reproductive tracts and the colon.[2][8][9] Epithelial sodium channels facilitate Na⁺ reabsorption across the apical membranes of epithelia in the distal nephron, respiratory and reproductive tracts and exocrine glands. Since Na⁺ ion concentration is a major determinant of extracellular fluid osmolarity, changes in Na⁺ concentration affect the movement of fluids and consequently fluid volume and blood pressure. The activity of ENaC in the colon and kidney is modulated by the mineralocorticoid aldosterone. It can be blocked by either triamterene or amiloride, which are used medically to serve as diuretics. In the kidney, it is inhibited by atrial natriuretic peptide, causing natriuresis and diuresis.
Epithelial Na+ channels (ENaCs) in the brain play a significant role in the regulation of blood pressure.[10] Vasopressin (VP) neurons play a pivotal role in coordinating neuroendocrine and autonomic responses to maintain cardiovascular homeostasis. High dietary salt intake causes an increase in the expression and activity of ENaC which results in the steady state depolarization of VP neurons.[10] This is one of the mechanisms underlying how dietary salt intake affects the activity of VP neurons via ENaC activity. ENaC channels in the brain are involved in blood pressure response to dietary sodium.
High-resolution immunofluorescence studies revealed that in the respiratory tract and the female reproductive tract, ENaC is located along the entire length of cilia that cover the surface of multi-ciliated cells.[8] Hence, in these epithelia with motile cilia, ENaC functions as a regulator of the osmolarity of the periciliary fluid, and its function is essential to maintain fluid volume at a depth necessary for the motility of the cilia. In the respiratory tract this movement is essential for clearing mucosal surface, and in the female reproductive tract, motility of the cilia is essential for the movement of oocytes.[8]
In contrast to ENaC, CFTR that regulates chloride ion transport is not found on cilia. These findings contradict a previous hypothesis that ENaC is downregulated by direct interaction with CFTR. In patients with cystic fibrosis (CF), CFTR cannot downregulate ENaC, causing hyper-absorption in the lungs and recurrent lung infections. It has been suggested that it may be a ligand-gated ion channel.[11]
In the skin epidermal layers, ENaC is expressed in the keratinocytes, sebaceous glands, and smooth muscle cells.[7] In these cells ENaC is mostly located in the cytoplasm.[7] In the eccrine sweat glands, ENaC is predominantly located in the apical membrane facing the lumen of the sweat ducts.[7] The major function of ENaC in these ducts is the re-uptake of Na⁺ ions that are excreted in sweat. In patients with ENaC mutations that cause systemic pseudohypoaldosteronism type I, the patients can lose a significant amount of Na⁺ ions, especially under hot climates.
ENaC is also found in taste receptors, where it plays an important role in saltiness perception. In rodents, virtually the entire salt taste is mediated by ENaC, whereas it seems to play a less significant role in humans: About 20 percent can be accredited to the epithelial sodium channel.
Protoelyzed variants of ENaC also function as human salt taste receptors. This role was first confirmed using human sensory studies to evaluate the effect of 4-propylphenyl 2-furoate on the perception of the salty taste of table salt, sodium chloride (NaCl). 4-propylphenyl 2-furoate is a compound that was discovered to activate proteolyzed ENaC.[12]
Ion selectivity
Studies show that the ENaC channel is permeable to Na+ and Li+ ions, but has very little permeability to K+, Cs+ or Rb+ ions.[13][14]
Transport reaction
The generalized transport reaction for Na+ channels is:
- Na+ (out) → Na+ (in)
That for the degenerins is:
- Cation (out) → cation (in)
Structure
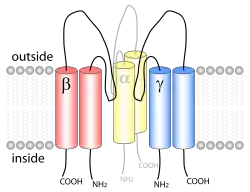
ENaC consists of three different subunits: α, β, γ.[2][15] All three subunits are essential for transport to the membrane assembly of functional channels on the membrane.[16] The C-terminus of each ENaC subunit contains a PPXY motif which when mutated or deleted in either the β- or γ-ENaC subunit leads to Liddle's syndrome, a human autosomal dominant form of hypertension. The cryoEM structure of ENaC indicates that the channel is a heterotrimeric protein like the acid-sensing ion channel 1 (ASIC1), which belongs to the same family.[17][18] Each of the subunits consists of two transmembrane helices and an extracellular loop. The amino- and carboxy-termini of all three polypeptides are located in the cytosol.
Crystal structure of ASIC1 and site-directed mutagenesis studies suggest that ENaC has a central ion channel located along the central symmetry axis in between the three subunits.[14][19]
In terms of structure, the proteins that belong to this family consist of about 510 to 920 amino acid residues. They are made of an intracellular N-terminus region followed by a transmembrane domain, a large extracellular loop, a second transmembrane segment, and a C-terminal intracellular tail.[20]
δ-subunit
In addition there is a fourth, so-called δ-subunit, that shares considerable sequence similarity with the α-subunit and can form a functional ion-channel together with the β- and γ-subunits. Such δ-, β-, γ-ENaC appear in pancreas, testes, lung, and ovaries. Their function is yet unknown.
Families
Members of the epithelial Na+ channel (ENaC) family fall into four subfamilies, termed alpha, beta, gamma and delta.[5] The proteins exhibit the same apparent topology, each with two transmembrane (TM)-spanning segments (TMS), separated by a large extracellular loop. In most ENaC proteins studied to date, the extracellular domains are highly conserved and contain numerous cysteine residues, with flanking C-terminal amphipathic TM regions, postulated to contribute to the formation of the hydrophilic pores of the oligomeric channel protein complexes. It is thought that the well-conserved extracellular domains serve as receptors to control the activities of the channels.
The vertebrate ENaC proteins from epithelial cells cluster tightly together on the phylogenetic tree; voltage-insensitive ENaC homologues are also found in the brain. The many sequenced C. elegans proteins, including the worm degenerins, are distantly related to the vertebrate proteins as well as to each other. Vertebrate ENaC proteins are similar to degenerins of Caenorhabditis elegans:[20] deg-1, del-1, mec-4, mec-10 and unc-8. These proteins can be mutated to cause neuronal degradation, and are also thought to form sodium channels.
Superfamily
The epithelial sodium (Na+) channel (ENaC) family belongs to the ENaC/P2X superfamily.[21] ENaC and P2X receptors have similar 3-d structures and are homologous.[22]
Genes
The exon–intron architecture of the three genes encoding the three subunits of ENaC have remained highly conserved despite the divergence of their sequences.[23]
There are four related amiloride sensitive sodium channels:
Stable ENaC Cell Line
Expression of ENaC in mammalian cell cultures is cytotoxic, resulting in sodium uptake, cell swelling and cell death, complicating production of stable cell lines to study ENaC. Chromovert technology enabled the production of a stable ENaC cell line using fluorogenic signaling probes and flow cytometry to scan numerous cells to isolate rare clones capable of functional, stable and viable expression of ENaC.[24]
Clinical significance

ENaC interaction with CFTR is of important pathophysiological relevance in cystic fibrosis. CFTR is a transmembrane channel responsible for chloride transport and defects in this protein cause cystic fibrosis, partly through upregulation of the ENaC channel in the absence of functional CFTR.
In the airways, CFTR allows for the secretion of chloride, and sodium ions and water follow passively. However, in the absence of functional CFTR, the ENaC channel is upregulated, and further decreases salt and water secretion by reabsorbing sodium ions. As such, the respiratory complications in cystic fibrosis are not solely caused by the lack of chloride secretion but instead by the increase in sodium and water reabsorption. This results in the deposition of thick, dehydrated mucus, which collects in the respiratory tract, interfering with gas exchange and allowing for the collection of bacteria.[25] Nevertheless, an upregulation of CFTR does not correct the influence of high-activity ENaC.[26] Probably other interacting proteins are necessary to maintain a functional ion homeostasis in epithelial tissue of the lung, like potassium channels, aquaporins or Na/K-ATPase.[27]
In sweat glands, CFTR is responsible for the reabsorption of chloride in the sweat duct. Sodium ions follow passively through ENaC as a result of the electrochemical gradient caused by chloride flow. This reduces salt and water loss. In the absence of chloride flow in cystic fibrosis, sodium ions do not flow through ENaC, leading to greater salt and water loss. (This is true despite upregulation of the ENaC channel, as flow in the sweat ducts is limited by the electrochemical gradient set up by chloride flow through CFTR.) As such, patients' skin tastes salty, and this is commonly used to help diagnose the disease, both in the past and today by modern electrical tests.[28]
Gain of function mutations to the β and γ subunits are associated with Liddle's syndrome.[29]
Amiloride and triamterene are potassium-sparing diuretics that act as epithelial sodium channel blockers.
References
- ↑ Noreng S, Bharadwaj A, Posert R, Yoshioka C, Baconguis I (September 2018). "Structure of the human epithelial sodium channel by cryo-electron microscopy". eLife. 7: e39340. doi:10.7554/eLife.39340. PMC 6197857. PMID 30251954.
- 1 2 3 4 Hanukoglu I, Hanukoglu A (April 2016). "Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases". Gene. 579 (2): 95–132. doi:10.1016/j.gene.2015.12.061. PMC 4756657. PMID 26772908.
- ↑ Shekdar K, Langer J, Venkatachalan S, Schmid L, Anobile J, Shah P, et al. (March 2021). "Cell engineering method using fluorogenic oligonucleotide signaling probes and flow cytometry". Biotechnology Letters. 43 (5): 949–958. doi:10.1007/s10529-021-03101-5. PMC 7937778. PMID 33683511.
- 1 2 Garty H (May 1994). "Molecular properties of epithelial, amiloride-blockable Na+ channels". FASEB Journal. 8 (8): 522–8. doi:10.1096/fasebj.8.8.8181670. PMID 8181670. S2CID 173677.
- 1 2 Le T, Saier MH (1996). "Phylogenetic characterization of the epithelial Na+ channel (ENaC) family". Molecular Membrane Biology. 13 (3): 149–57. doi:10.3109/09687689609160591. PMID 8905643.
- ↑ Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M (November 1995). "Molecular cloning and functional expression of a novel amiloride-sensitive Na+ channel". The Journal of Biological Chemistry. 270 (46): 27411–4. doi:10.1074/jbc.270.46.27411. PMID 7499195.
- 1 2 3 4 Hanukoglu I, Boggula VR, Vaknine H, Sharma S, Kleyman T, Hanukoglu A (June 2017). "Expression of epithelial sodium channel (ENaC) and CFTR in the human epidermis and epidermal appendages". Histochemistry and Cell Biology. 147 (6): 733–748. doi:10.1007/s00418-016-1535-3. PMID 28130590. S2CID 8504408.
- 1 2 3 Enuka Y, Hanukoglu I, Edelheit O, Vaknine H, Hanukoglu A (March 2012). "Epithelial sodium channels (ENaC) are uniformly distributed on motile cilia in the oviduct and the respiratory airways". Histochemistry and Cell Biology. 137 (3): 339–53. doi:10.1007/s00418-011-0904-1. PMID 22207244. S2CID 15178940.
- ↑ Sharma S, Hanukoglu A, Hanukoglu I (April 2018). "Localization of epithelial sodium channel (ENaC) and CFTR in the germinal epithelium of the testis, Sertoli cells, and spermatozoa". Journal of Molecular Histology. 49 (2): 195–208. doi:10.1007/s10735-018-9759-2. PMID 29453757. S2CID 3761720.
- 1 2 Sharma K, Haque M, Guidry R, Ueta Y, Teruyama R (September 2017). "Effect of dietary salt intake on epithelial Na+ channels (ENaC) in vasopressin magnocellular neurosecretory neurons in the rat supraoptic nucleus". The Journal of Physiology. 595 (17): 5857–5874. doi:10.1113/JP274856. PMC 5577521. PMID 28714095.
- ↑ Horisberger JD, Chraïbi A (2004). "Epithelial sodium channel: a ligand-gated channel?". Nephron Physiology. 96 (2): 37–41. doi:10.1159/000076406. PMID 14988660. S2CID 24608792.
- ↑ Shekdar K, Langer J, Venkatachalan S, Schmid L, Anobile J, Shah P, et al. (March 2021). "Cell engineering method using fluorogenic oligonucleotide signaling probes and flow cytometry". Biotechnology Letters. 43 (5): 949–958. doi:10.1007/s10529-021-03101-5. PMC 7937778. PMID 33683511.
- ↑ Happle R (October 1990). "Ptychotropism as a cutaneous feature of the CHILD syndrome". Journal of the American Academy of Dermatology. 23 (4 Pt 1): 763–6. doi:10.1016/0190-9622(90)70285-p. PMID 2229513.
- 1 2 Hanukoglu I (February 2017). "ASIC and ENaC type sodium channels: conformational states and the structures of the ion selectivity filters". The FEBS Journal. 284 (4): 525–545. doi:10.1111/febs.13840. PMID 27580245. S2CID 24402104.
- ↑ Loffing J, Schild L (November 2005). "Functional domains of the epithelial sodium channel". Journal of the American Society of Nephrology. 16 (11): 3175–81. doi:10.1681/ASN.2005050456. PMID 16192417.
- ↑ Edelheit O, Hanukoglu I, Dascal N, Hanukoglu A (April 2011). "Identification of the roles of conserved charged residues in the extracellular domain of an epithelial sodium channel (ENaC) subunit by alanine mutagenesis". American Journal of Physiology. Renal Physiology. 300 (4): F887-97. doi:10.1152/ajprenal.00648.2010. PMID 21209000. S2CID 869654.
- ↑ Noreng S, Bharadwaj A, Posert R, Yoshioka C, Baconguis I (September 2018). "Structure of the human epithelial sodium channel by cryo-electron microscopy". eLife. 7: e39340. doi:10.7554/eLife.39340. PMC 6197857. PMID 30251954.
- ↑ Jasti J, Furukawa H, Gonzales EB, Gouaux E (September 2007). "Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH". Nature. 449 (7160): 316–23. Bibcode:2007Natur.449..316J. doi:10.1038/nature06163. PMID 17882215.
- ↑ Edelheit O, Ben-Shahar R, Dascal N, Hanukoglu A, Hanukoglu I (April 2014). "Conserved charged residues at the surface and interface of epithelial sodium channel subunits--roles in cell surface expression and the sodium self-inhibition response". The FEBS Journal. 281 (8): 2097–111. doi:10.1111/febs.12765. PMID 24571549. S2CID 5807500.
- 1 2 Snyder PM, McDonald FJ, Stokes JB, Welsh MJ (September 1994). "Membrane topology of the amiloride-sensitive epithelial sodium channel". The Journal of Biological Chemistry. 269 (39): 24379–83. doi:10.1016/S0021-9258(19)51094-8. PMID 7929098.
- ↑ "ATP-gated P2X Receptor Cation Channel (P2X Receptor) Family". Functional and Phylogenetic Classification of Membrane Transport Proteins. Saier Lab. Group, UCSD and SDSC.
- ↑ Chen JS, Reddy V, Chen JH, Shlykov MA, Zheng WH, Cho J, et al. (2011). "Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments". Journal of Molecular Microbiology and Biotechnology. 21 (3–4): 83–96. doi:10.1159/000334611. PMC 3290041. PMID 22286036.
- ↑ Saxena A, Hanukoglu I, Strautnieks SS, Thompson RJ, Gardiner RM, Hanukoglu A (November 1998). "Gene structure of the human amiloride-sensitive epithelial sodium channel beta subunit". Biochemical and Biophysical Research Communications. 252 (1): 208–13. doi:10.1006/bbrc.1998.9625. PMID 9813171.
- ↑ Shekdar K, Langer J, Venkatachalan S, Schmid L, Anobile J, Shah P, et al. (March 2021). "Cell engineering method using fluorogenic oligonucleotide signaling probes and flow cytometry". Biotechnology Letters. 43 (5): 949–958. doi:10.1007/s10529-021-03101-5. PMC 7937778. PMID 33683511.
- ↑ Mall M, Grubb BR, Harkema JR, O'Neal WK, Boucher RC (May 2004). "Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice". Nature Medicine. 10 (5): 487–93. doi:10.1038/nm1028. PMID 15077107. S2CID 45366866.
- ↑ Grubb BR, O'Neal WK, Ostrowski LE, Kreda SM, Button B, Boucher RC (January 2012). "Transgenic hCFTR expression fails to correct β-ENaC mouse lung disease". American Journal of Physiology. Lung Cellular and Molecular Physiology. 302 (2): L238-47. doi:10.1152/ajplung.00083.2011. PMC 3349361. PMID 22003093.
- ↑ Toczyłowska-Mamińska R, Dołowy K (February 2012). "Ion transporting proteins of human bronchial epithelium". Journal of Cellular Biochemistry. 113 (2): 426–32. doi:10.1002/jcb.23393. PMID 21975871. S2CID 31970069.
- ↑ Berdiev BK, Qadri YJ, Benos DJ (February 2009). "Assessment of the CFTR and ENaC association". Molecular BioSystems. 5 (2): 123–7. doi:10.1039/B810471A. PMC 2666849. PMID 19156256.
- ↑ Ion Channel Diseases
External links
- Epithelial+sodium+channel at the U.S. National Library of Medicine Medical Subject Headings (MeSH)