Artificial transcription factors (ATFs) are engineered individual or multi molecule transcription factors that either activate or repress gene transcription (biology).[1]
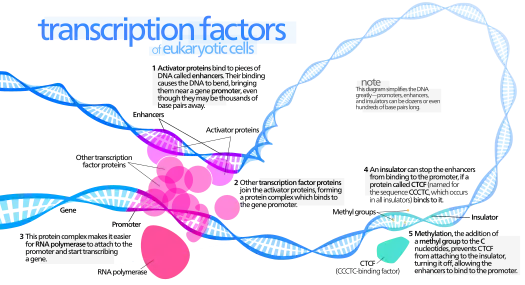
ATFs often contain two main components linked together, a DNA-binding domain and a regulatory domain, also known as an effector domain or modulatory domain.[1] The DNA-binding domain targets a specific DNA sequence with high affinity, and the regulatory domain is responsible for activating or repressing the bound gene.[1] The ATF can directly regulate gene expression, can recruit proteins and other transcription factors to initiate transcription, or recruit proteins and other transcription factors to compact the DNA which inhibits RNA polymerase from binding and transcribing the DNA; an example of transcription factors up-regulating gene expression is displayed in figure 1 on the left.[1][2] Because ATFs are composed of two separable components, the DNA-binding domain and the regulatory domain, the two domains are interchangeable, permitting the design of new ATFs from existing natural transcription factors.[1]
Some applications of ATFs include reprogramming cell state, cancer treatment, and a plausible treatment for Angelman Syndrome.[2][3][4]
ATF Design
DNA-Binding Domain
The DNA-binding domain routes the ATF to a specific gene sequence. Natural DNA binding proteins are commonly used because of their high affinity for their DNA target sequence, however currently no algorithm that matches the protein amino-acid sequence to the complementary DNA binding sequence exists, limiting the rational design of new DNA-binding proteins.[1] Non-peptide, oligonucleotide, and polyamide DNA-binding domains have recently been explored which permit rational design.[1] The type of DNA binding domain chosen depends on the desired application of the ATF, common DNA-binding domains are presented in Types of ATF DNA-Binding Domains section below.[1][2]
Regulatory Domain
The regulatory domain is responsible for activating or repressing the bound gene and accomplishes this regulation by either directly regulating gene expression or recruiting other proteins and transcription factors to change transcription levels.[1][2] One route to upregulate a gene is for the ATF to recruit proteins that loosen the DNA wrapping around histones allowing RNA polymerase to bind and transcribe the gene; likewise, compacting the DNA would downregulate gene expression by inhibiting RNA polymerase from binding.[1] Regulatory domains promoting gene transcription are usually acidic activators, composed of acidic and hydrophobic amino acids, and regulatory domains repressing gene transcription usually contain more basic amino acids.[1] Factors influencing the effect the ATF has on transcription include the distance the regulatory domain is from the transcription site, the cell type, and the number of activating or repressing sequences present in the regulatory domain.[1] Activating domains, regulatory domains that promote gene transcription, are often capable of upregulating transcription by 5 to 40-fold and RNA regulatory domains have been shown to result in 100 fold transcription levels.[1] An alternative strategy for repressing genes is for the ATF to out-compete natural transcriptions factors and physically block transcription by RNA polymerase; however, creating ATFs with higher affinity for the DNA sequence than the natural transcription factors remains a challenge.[1]
Linkers
Linkers covalently or non-covalently link the DNA-binding domain and regulatory domain.[1] Frequently, peptide linkers are used, but polyethylene glycol and small molecules linkers also exist.[1] The linkers enable the DNA-binding domains and regulatory domains to be interchangeable allowing the design of new ATFs from natural transcription factor components.[1] Although linkers are less studied, the linker length is important because it alters the extent of impact the regulatory domain has on gene expression.[1]
History
Most ATFs have been constructed by exchanging existing DNA-binding domains and regulatory domains to generate ATFs with new targeting sites and transcription regulation consequences.[1] Designed DNA-binding domains, such as CRISPR-Cas, with new targeting capabilities are being explored to engineer higher specificity and control potential side effects.[2] In the future, ATFs which can respond to physiological cues, only change transcription levels in a specific cell type, and can easily be delivered without the use of electroporation are of great interest.[1]
Types of ATF DNA-Binding Domains
CRISPR-Cas
The clustered regularly interspaced short palindromic repeats - Cas (CRISPR-Cas) system has been extensively studied to target a specific DNA sequence using a single guide RNA (sgRNA).[5] For ATF applications the CRISPR-Cas system is modified to inactivate the Cas enzyme's natural function and link a regulatory domain to the Cas enzyme.[2] The CRISPR-Cas system benefits from high specificity between the sgRNA and the target DNA sequence and the simplicity of designing new sgRNAs; however, the CRISPR-Cas system requires a PAM sequence directly upstream of the target DNA site and the large size of the Cas protein hinders delivery into the cell.[2]
TALEs
Transcription activator-like effectors (TALEs) are peptide structures composed of repeating 34 amino acids long segments forming a peptide ranging in total length from 340 to 510 amino acids.[2] Each repeating segment folds into two alpha helices and amino acids at residue positions 12 and 13 in the repeating segment determines the DNA binding sequence.[2] The TALEs peptide has high specificity to the target DNA preventing secondary side effects, but this high specificity prevents the ATF from binding to multiple sites and requires a different ATF for each desired effect.[2]
Zinc Fingers
Zinc fingers are naturally abundant, involved in multiple regulatory processes, and are common eukaryotic transcriptional factors.[6] Cis2/His2 zinc fingers have been extensively studied, are composed of 30 amino acids, can bind to non-palindromic sequences, and contain 3 to 4 critical amino acids at positions 1, 3, and 6 on the alpha helix which designate the complementary binding sequence.[4][7][8] Because zinc fingers are only 30 amino acids long they are easier to deliver, and multiple zinc fingers can be linked together to target larger DNA sequences with one ATF; however, connecting more than three zinc fingers together reduces each zinc finger’s specificity and increases off-site targeting.[2]
ATF Applications
Reprogramming Cell State
Directing cell differentiation and reprogramming cell fate have traditionally been achieved via a mixture of transcription factors.[9] The field gained significant interest once four transcription factors Oct4/Sox2/cMyc/Klf4 were found to reprogram cells from a differentiated state into an induced pluripotent stem cell state similar to embryonic stem cells.[10] Multiple ATFs composed of three zinc finger proteins linked together can each activate genes that eventually lead to the production of the Oct4 transcription factor in the cell, causing the cell to reprogram to an induced pluripotent state without the addition of external Oct4 transcription factors.[2] The change in cell state demonstrates that ATFs can replace traditional transcription factors in cell reprogramming.[2]
Angelman Syndrome
Angelman syndrome is a neurological development disorder caused by the deactivation of the maternal UBE3A gene.[3] Two potential treatment strategies using ATFs are to upregulate the expression of the maternal UBE3A gene or downregulate the expression of UBE3A-AS gene, the gene that causes repression of the paternal UBE3A gene.[3] Zinc finger ATF TAT-S1 acts as a strong repressor against the UBE3A-AS gene, and when administered to mice, resulted in increased Ube3a in the brain.[3]
Cancer
Abnormal gene expression is regularly associated with cancer and uncontrolled tumor growth, making ATFs a promising therapeutic for cancer treatment.[4] By linking 6 zinc fingers together in an ATF, the ATF only binds to an 18 base pair sequence containing smaller subsequences complementary to each zinc finger in the ATF, so the ATF is more specific than one zinc finger which only targets a specific 3 to 4 base pair sequence.[4] ATFs linked to the KRAB repressor regulatory domain decreases cancer cells' drug resistance to chemotherapy, and ATFs linked to activator domains can upregulate Bax gene expression causing cell apoptosis; however, these treatments remain in the early stages because of inadequate delivery methods.[4]
See also
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Ansari, Aseem Z; Mapp, Anna K (2002-12-01). "Modular design of artificial transcription factors". Current Opinion in Chemical Biology. 6 (6): 765–772. doi:10.1016/S1367-5931(02)00377-0. ISSN 1367-5931. PMID 12470729.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Heiderscheit, Evan A.; Eguchi, Asuka; Spurgat, Mackenzie C.; Ansari, Aseem Z. (2018). "Reprogramming cell fate with artificial transcription factors". FEBS Letters. 592 (6): 888–900. doi:10.1002/1873-3468.12993. ISSN 1873-3468. PMC 5869137. PMID 29389011.
- 1 2 3 4 Tan, Wen-Hann; Bird, Lynne M. (December 2016). "Angelman syndrome: Current and emerging therapies in 2016". American Journal of Medical Genetics. Part C, Seminars in Medical Genetics. 172 (4): 384–401. doi:10.1002/ajmg.c.31536. ISSN 1552-4876. PMID 27860204. S2CID 4377191.
- 1 2 3 4 5 Yan, Chunhong; Higgins, Paul J. (2013-01-01). "Drugging the undruggable: Transcription therapy for cancer". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1835 (1): 76–85. doi:10.1016/j.bbcan.2012.11.002. ISSN 0304-419X. PMC 3529832. PMID 23147197.
- ↑ Nidhi, Sweta; Anand, Uttpal; Oleksak, Patrik; Tripathi, Pooja; Lal, Jonathan A.; Thomas, George; Kuca, Kamil; Tripathi, Vijay (2021-03-24). "Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives". International Journal of Molecular Sciences. 22 (7): 3327. doi:10.3390/ijms22073327. ISSN 1422-0067. PMC 8036902. PMID 33805113.
- ↑ Cassandri, Matteo; Smirnov, Artem; Novelli, Flavia; Pitolli, Consuelo; Agostini, Massimiliano; Malewicz, Michal; Melino, Gerry; Raschellà, Giuseppe (2017-11-13). "Zinc-finger proteins in health and disease". Cell Death Discovery. 3 (1): 17071. doi:10.1038/cddiscovery.2017.71. ISSN 2058-7716. PMC 5683310. PMID 29152378.
- ↑ Gommans, Willemijn M.; Haisma, Hidde J.; Rots, Marianne G. (2005-12-02). "Engineering Zinc Finger Protein Transcription Factors: The Therapeutic Relevance of Switching Endogenous Gene Expression On or Off at Command". Journal of Molecular Biology. 354 (3): 507–519. doi:10.1016/j.jmb.2005.06.082. ISSN 0022-2836. PMID 16253273.
- ↑ Urnov, Fyodor D; Rebar, Edward J (2002-09-01). "Designed transcription factors as tools for therapeutics and functional genomics". Biochemical Pharmacology. Cell Signaling, Transcription and Translation as Therapeutic Targets. 64 (5): 919–923. doi:10.1016/S0006-2952(02)01150-4. ISSN 0006-2952. PMID 12213587.
- ↑ Takahashi, Kazutoshi; Yamanaka, Shinya (March 2016). "A decade of transcription factor-mediated reprogramming to pluripotency". Nature Reviews Molecular Cell Biology. 17 (3): 183–193. doi:10.1038/nrm.2016.8. ISSN 1471-0080. PMID 26883003. S2CID 7593915.
- ↑ Qi, Huayu; Pei, Duanqing (July 2007). "The magic of four: induction of pluripotent stem cells from somatic cells by Oct4, Sox2, Myc and Klf4". Cell Research. 17 (7): 578–580. doi:10.1038/cr.2007.59. ISSN 1748-7838. PMID 17632550. S2CID 9643825.