| L-threonine ammonia-lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 A 3d cartoon depiction of the threonine deaminase tetramer | |||||||||
| Identifiers | |||||||||
| EC no. | 4.3.1.19 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Threonine ammonia-lyase (EC 4.3.1.19, systematic name L-threonine ammonia-lyase (2-oxobutanoate-forming), also commonly referred to as threonine deaminase or threonine dehydratase, is an enzyme responsible for catalyzing the conversion of L-threonine into α-ketobutyrate and ammonia:
- L-threonine = 2-oxobutanoate + NH3 (overall reaction)
- (1a) L-threonine = 2-aminobut-2-enoate + H2O
- (1b) 2-aminobut-2-enoate = 2-iminobutanoate (spontaneous)
- (1c) 2-iminobutanoate + H2O = 2-oxobutanoate + NH3 (spontaneous)
α-Ketobutyrate can be converted into L-isoleucine, so threonine ammonia-lyase functions as a key enzyme in BCAA synthesis.[1] It employs a pyridoxal-5'-phosphate cofactor, similar to many enzymes involved in amino acid metabolism. It is found in bacteria, yeast, and plants, though most research to date has focused on forms of the enzyme in bacteria. This enzyme was one of the first in which negative feedback inhibition by the end product of a metabolic pathway was directly observed and studied.[2] The enzyme serves as an excellent example of the regulatory strategies used in amino acid homeostasis.
Structure
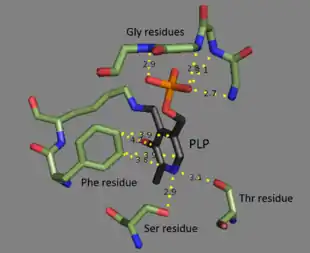
Threonine ammonia-lyase is a tetramer of identical subunits, and is arranged as a dimer of dimers. Each subunit has two domains: a domain containing the catalytic active site and a domain with allosteric regulatory sites. The two have been shown to be distinct regions,[3] but the regulatory site of one subunit actually interacts with the catalytic site of another subunit.[4] Both domains contain the repeating structural motif of beta sheets surrounded by alpha helices.[5] While the threonine binding site is not perfectly understood, structural studies do reveal how the pyridoxal phosphate cofactor is bound.[4] The PLP cofactor is bonded to a lysine residue by means of a Schiff base, and the phosphate group of PLP is held in place by amine groups derived from a repeating sequence of glycine residues. The aromatic ring is bound to phenylalanine, and the nitrogen on the ring is hydrogen bonded to hydroxyl group-containing residues.[6]
Mechanism
The mechanism of threonine ammonia-lyase is analogous to other deaminating PLP enzymes in its use of Schiff base intermediates.[7] Initially, the amine group of threonine attacks the lysine/PLP Schiff base, displacing lysine. After deprotonation of the amino acid alpha carbon and subsequent dehydration (hence the common name threonine dehydratase), a new Schiff base is formed. This Schiff base is replaced by lysine attack, reforming the catalytically active PLP and releasing an initial alkene-containing product. This product tautomerizes, and after hydrolysis of the Schiff base, the final products are generated.[8][9] After the final alpha-ketobutyrate product is generated, isoleucine is synthesized by progressing through the intermediates alpha-acetohydroxybutyrate to alpha-beta-dihydroxy-beta-methylvalerate, then to alpha-keto-beta-methylvalerate.[10]
Regulation
Threonine ammonia-lyase has been shown to not follow Michaelis-Menten kinetics, rather, it is subject to complex allosteric control.[11] The enzyme is inhibited by isoleucine, the product of the pathway it participates in, and is activated by valine, the product of a parallel pathway.[1] Thus, an increase in isoleucine concentration shuts off its production, and an increase in valine concentration diverts starting material (Hydroxyethyl-TPP) away from valine production. The enzyme has two binding sites for isoleucine; one has a high affinity for isoleucine and the other has a low affinity.[12] The binding of isoleucine to the high affinity site increases the binding affinity of the low affinity site, and enzyme deactivation occurs when isoleucine binds to the low affinity site. Valine promotes enzyme activity by competitively binding to the high affinity site, preventing isoleucine from having an inhibitory effect.[12] The combination of these two feedback methods balances the concentration of BCAAs.

Isoforms and other functions
Multiple forms of threonine ammonia-lyase have been observed in a variety of species of organism. In Escherichia coli, a system in which the enzyme has been studied extensively, two different forms of the enzyme are found. One is biosynthetic and resembles the enzyme characteristics presented here, while the other is degradative and functions to generate carbon fragments for energy production.[2] The pair of isoforms has also been observed in other bacteria. In many bacteria, the biodegradative isoform of the enzyme is expressed in anaerobic conditions and is promoted by cAMP and threonine, while the biosynthetic isoform is expressed in aerobic conditions.[13] This allows the bacterium to balance energy stores and inhibit energy-consuming synthetic pathways when energy is not abundant.
In plants, threonine ammonia-lyase is important in defense mechanisms against herbivores and is upregulated in response to abiotic stress.[14] An adapted isoform of the enzyme with unique properties that deter herbivores is expressed in plant leaves. The catalytic domain of this isoform is extremely resistant to proteolysis, while the regulatory domain degrades readily, so upon ingestion by another organism, the threonine deamination capabilities of the enzyme go unchecked. This degrades threonine before the herbivore can absorb it, starving the herbivore of an essential amino acid.[15] Studies of threonine ammonia-lyase in plants have also offered new strategies in the development of GMOs with increased nutritional value by increasing essential amino acid content.[14]
Other more exotic forms of the enzyme have been found that are extremely small in size, but still retain all catalytic and regulatory functions.[4]
Evolution
There are five major fold types for PLP-dependent enzymes. Threonine ammonia-lyase is a member of the Fold Type II family, also known as the tryptophan synthase family.[7] Though threonine ammonia-lyase does not possess substrate tunneling like tryptophan synthase does, it contains much conserved homology. Threonine ammonia-lyase is most closely related to serine dehydratase, and both possess the same general catalytic mechanism.[9] In fact, threonine ammonia-lyase has been shown to exhibit some specificity towards serine and can convert serine into pyruvate.[2] The regulatory domain of threonine ammonia-lyase is very similar to the regulatory domain of phosphoglycerate dehydrogenase.[4] All of these relationships demonstrate that threonine ammonia-lyase has close evolutionary ties to these enzymes. Due to the degree of conserved structure and sequence in enzymes that recognize amino acids, it is likely that the evolutionary diversity of these enzymes came about by the matching together of individual regulatory and catalytic domains in various ways.[1]
Relevance to humans
Threonine ammonia-lyase is not found in humans. Thus, this is one example of why humans cannot synthesize all 20 proteinogenic amino acids; in this specific case, humans cannot convert threonine into isoleucine and must consume isoleucine in the diet.[1] The enzyme has also been studied in the past as a possible tumor suppressing agent for the previously described reasons, in that it deprives tumor cells of an essential amino acid and kills them,[16] but this treatment has not been utilized.
References
- 1 2 3 4 5 Berg JM, Tymoczko JL, Stryer L (2012). Biochemistry (7th ed.). New York: W.H. Freeman and Company. ISBN 978-1-4292-7635-1.
- 1 2 3 Umbarger HE, Brown B (January 1957). "Threonine deamination in Escherichia coli. II. Evidence for two L-threonine deaminases". Journal of Bacteriology. 73 (1): 105–12. doi:10.1128/jb.73.1.105-112.1957. PMC 289754. PMID 13405870.
- ↑ Changeux J (1963). "Allosteric Interactions on Biosynthetic L-threonine Deaminase from E. coli K12". Cold Spring Harbor Symposia on Quantitative Biology. 28: 497–504. doi:10.1101/SQB.1963.028.01.066.
- 1 2 3 4 Gallagher DT, Gilliland GL, Xiao G, Zondlo J, Fisher KE, Chinchilla D, Eisenstein E (April 1998). "Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase". Structure. 6 (4): 465–75. doi:10.1016/s0969-2126(98)00048-3. PMID 9562556.
- ↑ Schneider G, Käck H, Lindqvist Y (January 2000). "The manifold of vitamin B6 dependent enzymes". Structure. 8 (1): R1-6. doi:10.1016/S0969-2126(00)00085-X. PMID 10673430.
- 1 2 Goto M (2005). "Crystal Structure of T.th. HB8 Threonine deaminase". doi:10.2210/pdb1ve5/pdb.
{{cite journal}}: Cite journal requires|journal=(help) - 1 2 Eliot AC, Kirsch JF (2004). "Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations". Annual Review of Biochemistry. 73 (1): 383–415. doi:10.1146/annurev.biochem.73.011303.074021. PMID 15189147.
- 1 2 Umbarger HE (2009). "Threonine Deaminases". In Meister A (ed.). Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology - and Related Areas of Molecular Biology. Vol. 37. John Wiley & Sons. pp. 349–95. doi:10.1002/9780470122822.ch6. ISBN 978-0-471-59172-6. PMID 4570068.
- 1 2 3 Jin J, Hanefeld U (March 2011). "The selective addition of water to C=C bonds; enzymes are the best chemists". Chemical Communications. 47 (9): 2502–10. doi:10.1039/c0cc04153j. PMID 21243161.
- ↑ Squires CH, Levinthal M, De Felice M (November 1981). "A role for threonine deaminase in the regulation of alpha-acetolactate biosynthesis in Escherichia coli K12". Journal of General Microbiology. 127 (1): 19–25. doi:10.1099/00221287-127-1-19. PMID 7040602.
- ↑ Changeux JP (1961). "The feedback control mechanisms of biosynthetic L-threonine deaminase by L-isoleucine". Cold Spring Harbor Symposia on Quantitative Biology. 26: 313–8. doi:10.1101/SQB.1961.026.01.037. PMID 13878122.
- 1 2 Wessel PM, Graciet E, Douce R, Dumas R (December 2000). "Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments" (PDF). Biochemistry. 39 (49): 15136–43. doi:10.1021/bi001625c. PMID 11106492.
- ↑ Luginbuhl GH, Hofler JG, Decedue CJ, Burns RO (October 1974). "Biodegradative L-threonine deaminase of Salmonella typhimurium". Journal of Bacteriology. 120 (1): 559–61. doi:10.1128/jb.120.1.559-561.1974. PMC 245803. PMID 4370904.
- 1 2 Joshi V, Joung JG, Fei Z, Jander G (October 2010). "Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress". Amino Acids. 39 (4): 933–47. doi:10.1007/s00726-010-0505-7. PMID 20186554. S2CID 22641155.
- ↑ Gonzales-Vigil E, Bianchetti CM, Phillips GN, Howe GA (April 2011). "Adaptive evolution of threonine deaminase in plant defense against insect herbivores". Proceedings of the National Academy of Sciences of the United States of America. 108 (14): 5897–902. Bibcode:2011PNAS..108.5897G. doi:10.1073/pnas.1016157108. PMC 3078374. PMID 21436043.
- ↑ Greenfield RS, Wellner D (August 1977). "Effects of threonine deaminase on growth and viability of mammalian cells in tissue culture and its selective cytotoxicity toward leukemia cells". Cancer Research. 37 (8 Pt 1): 2523–9. PMID 559542.
