| ITPA | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ITPA, C20orf37, HLC14-06-P, dJ794I6.3, My049, ITPase, NTPase, inosine triphosphatase, DEE35 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 147520 MGI: 96622 HomoloGene: 6289 GeneCards: ITPA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Inosine triphosphate pyrophosphatase is an enzyme that in humans is encoded by the ITPA gene,[5][6] by the rdgB gene in bacteria E.coli[7] and the HAM1 gene in yeast S. cerevisiae;[8] the protein is also encoded by some RNA viruses of the Potyviridae family.[9] Two transcript variants encoding two different isoforms have been found for this gene. Also, at least two other transcript variants have been identified which are probably regulatory rather than protein-coding.
Function
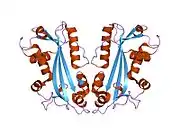
The protein encoded by this gene hydrolyzes inosine triphosphate and deoxyinosine triphosphate to the monophosphate nucleotide and diphosphate.[6] The enzyme possesses a multiple substrate-specificity and acts on other nucleotides including xanthosine triphosphate and deoxyxanthosine triphosphate.[8] The encoded protein, which is a member of the HAM1 NTPase protein family, is found in the cytoplasm and acts as a homodimer.
Clinical significance
Defects in the encoded protein can result in inosine triphosphate pyrophosphorylase deficiency.[6] The enzyme ITPase dephosphorylates ribavirin triphosphate in vitro to ribavirin monophosphate, and ITPase reduced enzymatic activity present in 30 % of humans potentiates mutagenesis in hepatitis C virus.[10] Gene variants predicting reduced predicted ITPase activity have been associated with decreased risk of ribavirin-induced anemia, increased risk of thrombocytopenia, lower ribavirin concentrations, as well as a ribavirin-like reduced relapse risk following interferon-based therapy for hepatitis C virus (HCV) genotype 2 or 3 infection. [11]
Reading
- Holmes SL, Turner BM, Hirschhorn K (1979). "Human inosine triphosphatase: catalytic properties and population studies". Clin. Chim. Acta. 97 (2–3): 143–53. doi:10.1016/0009-8981(79)90410-8. PMID 487601.
- Fraser JH, Meyers H, Henderson JF, et al. (1976). "Individual variation in inosine triphosphate accumulation in human erythrocytes". Clin. Biochem. 8 (6): 353–64. doi:10.1016/S0009-9120(75)93685-1. PMID 1204209.
- Clawson GA, Song YL, Schwartz AM, et al. (1992). "Interaction of human immunodeficiency virus type I Rev protein with nuclear scaffold nucleoside triphosphatase activity". Cell Growth Differ. 2 (11): 575–82. PMID 1667585.
- Deloukas P, Matthews LH, Ashurst J, et al. (2002). "The DNA sequence and comparative analysis of human chromosome 20". Nature. 414 (6866): 865–71. Bibcode:2001Natur.414..865D. doi:10.1038/414865a. PMID 11780052.
- Sumi S, Marinaki AM, Arenas M, et al. (2002). "Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency". Hum. Genet. 111 (4–5): 360–7. doi:10.1007/s00439-002-0798-z. PMID 12384777. S2CID 24240940.
- Cao H, Hegele RA (2003). "DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency". J. Hum. Genet. 47 (11): 620–2. doi:10.1007/s100380200095. PMID 12436200.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Marinaki AM, Duley JA, Arenas M, et al. (2005). "Mutation in the ITPA gene predicts intolerance to azathioprine". Nucleosides Nucleotides Nucleic Acids. 23 (8–9): 1393–7. doi:10.1081/NCN-200027639. PMID 15571265. S2CID 86308163.
- Marinaki AM, Sumi S, Arenas M, et al. (2005). "Allele frequency of inosine triphosphate pyrophosphatase gene polymorphisms in a Japanese population". Nucleosides Nucleotides Nucleic Acids. 23 (8–9): 1399–401. doi:10.1081/NCN-200027641. PMID 15571266. S2CID 36126159.
- Maeda T, Sumi S, Ueta A, et al. (2005). "Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency in the Japanese population". Mol. Genet. Metab. 85 (4): 271–9. doi:10.1016/j.ymgme.2005.03.011. PMID 15946879.
- Breen DP, Marinaki AM, Arenas M, Hayes PC (2005). "Pharmacogenetic association with adverse drug reactions to azathioprine immunosuppressive therapy following liver transplantation". Liver Transpl. 11 (7): 826–33. doi:10.1002/lt.20377. PMID 15973722. S2CID 9301778.
- Porta J, Kolar C, Kozmin SG, et al. (2006). "Structure of the orthorhombic form of human inosine triphosphate pyrophosphatase". Acta Crystallographica Section F. 62 (Pt 11): 1076–81. doi:10.1107/S1744309106041790. PMC 2225220. PMID 17077483.
- Arenas M, Duley J, Sumi S, et al. (2007). "The ITPA c.94C>A and g.IVS2+21A>C sequence variants contribute to missplicing of the ITPA gene". Biochim. Biophys. Acta. 1772 (1): 96–102. doi:10.1016/j.bbadis.2006.10.006. PMID 17113761.
- Stenmark P, Kursula P, Flodin S, et al. (2007). "Crystal structure of human inosine triphosphatase. Substrate binding and implication of the inosine triphosphatase deficiency mutation P32T". J. Biol. Chem. 282 (5): 3182–7. doi:10.1074/jbc.M609838200. PMID 17138556.
- Atanasova S, Shipkova M, Svinarov D, et al. (2007). "Analysis of ITPA phenotype-genotype correlation in the Bulgarian population revealed a novel gene variant in exon 6". Therapeutic Drug Monitoring. 29 (1): 6–10. doi:10.1097/FTD.0b013e3180308554. PMID 17304144. S2CID 7286658.
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000125877 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000074797 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ Lin S, McLennan AG, Ying K, Wang Z, Gu S, Jin H, Wu C, Liu W, Yuan Y, Tang R, Xie Y, Mao Y (May 2001). "Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene". J Biol Chem. 276 (22): 18695–701. doi:10.1074/jbc.M011084200. PMID 11278832.
- 1 2 3 "Entrez Gene: ITPA inosine triphosphatase (nucleoside triphosphate pyrophosphatase)".
- ↑ Burgis NE, Cunningham RP (2007). "Substrate specificity of RdgB protein, a deoxyribonucleoside triphosphate pyrophosphohydrolase". J Biol Chem. 282 (8): 3531–8. doi:10.1074/jbc.M608708200. PMID 17090528.
- 1 2 Davies O, Mendes P, Smallbone K, Malys N (2012). "Characterisation of multiple substrate-specific (d)ITP/(d)XTPase and modelling of deaminated purine nucleotide metabolism". BMB Reports. 45 (4): 259–64. doi:10.5483/BMBRep.2012.45.4.259. PMID 22531138.
- ↑ Pasin, Fabio; Daròs, José-Antonio; Tzanetakis, Ioannis E (2022). "Proteome expansion in the Potyviridae evolutionary radiation". FEMS Microbiology Reviews. 46 (4): fuac011. doi:10.1093/femsre/fuac011. ISSN 1574-6976. PMC 9249622. PMID 35195244.
- ↑ Nyström K, Wanrooij PH, Waldenström J, Adamek L, Brunet S, Said J, Nilsson S, Wind-Rotolo M, Hellstrand K, Norder H, Tang KW, Lagging M (October 2018). "Inosine Triphosphate Pyrophosphatase Dephosphorylates Ribavirin Triphosphate and Reduced Enzymatic Activity Potentiates Mutagenesis in Hepatitis C Virus". Journal of Virology. 92 (19): 01087–18. doi:10.1002/hep.27009. PMC 6146798. PMID 30045981.
- ↑ Rembeck K, Waldenstrom J, Hellstrand K, Nilsson S, Nyström K, Martner A, Lindh M, Norkrans G, Westin J, Pedersen C, Färkkil M, Langeland N, Rauning Buhl M, Morch K, Brehm Christensen P, and Lagging M (June 2014). "Variants of the inosine triphosphate pyrophosphatase gene are associated with reduced relapse risk following treatment for HCV genotype 2/3". Hepatology. 59 (6): 2131–2139. doi:10.1002/hep.27009. PMID 24519039.