In chemistry, dimerization refers to the process of joining two molecular entities by bonds. The resulting bonds can be either strong or weak. Many symmetrical chemical species are described as dimers, even when the monomer is unknown or highly unstable.[1]
The term homodimer is used when the two subunits are identical (e.g. A–A) and heterodimer when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely-charged ions associate into dimers, they are referred to as Bjerrum pairs,[2] after Danish chemist Niels Bjerrum.
Noncovalent dimers

Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds.[3] Many OH-containing molecules form dimers, e.g. the water dimer.
Excimers and exciplexes are excited structures with a short lifetime. For example, noble gases do not form stable dimers, but they do form the excimers Ar2*, Kr2* and Xe2* under high pressure and electrical stimulation.[4]
Covalent dimers


Molecular dimers are often formed by the reaction of two identical compounds e.g.: 2A → A−A. In this example, monomer "A" is said to dimerize to give the dimer "A−A". An example is a diaminocarbene, which dimerize to give a tetraaminoethylene:
Carbenes are highly reactive and readily form bonds.
Dicyclopentadiene is an asymmetrical dimer of two cyclopentadiene molecules that have reacted in a Diels-Alder reaction to give the product. Upon heating, it "cracks" (undergoes a retro-Diels-Alder reaction) to give identical monomers:
Many nonmetallic elements occur as dimers: hydrogen, nitrogen, oxygen, and the halogens (i.e. fluorine, chlorine, bromine and iodine). Noble gases can form dimers linked by van der Waals bonds, such as dihelium or diargon. Mercury occurs as a mercury(I) cation (Hg2+2), formally a dimeric ion. Other metals may form a proportion of dimers in their vapour phase. Known metallic dimers include dilithium (Li2), disodium (Na2), dipotassium (K2), dirubidium (Rb2) and dicaesium (Cs2). Such elemental dimers are homonuclear diatomic molecules.
Many small organic molecules, most notably formaldehyde, easily form dimers. The dimer of formaldehyde (CH2O) is dioxetane (C2H4O2).
Borane (BH3) occurs as the dimer diborane (B2H6), due to the high Lewis acidity of the boron center.
Polymer chemistry
In the context of polymers, "dimer" also refers to the degree of polymerization 2, regardless of the stoichiometry or condensation reactions.
One case where this is applicable is with disaccharides. For example, cellobiose is a dimer of glucose, even though the formation reaction produces water:
Here, the resulting dimer has a stoichiometry different from the initial pair of monomers.
Disaccharides need not be composed of the same monosaccharides to be considered dimers. An example is sucrose, a dimer of fructose and glucose, which follows the same reaction equation as presented above.
Amino acids can also form dimers, which are called dipeptides. An example is glycylglycine, consisting of two glycine molecules joined by a peptide bond. Other examples include aspartame and carnosine.
Inorganic dimers
Many molecules and ions are described as dimers, even when the monomer is elusive.
Group 13 dimers
Boranes

Diborane (B2H6) is an inorganic dimer of Borane. B2H6 exists as a structure where two hydrogen atoms bridge the two boron atoms.[5]
Aluminium

Trialkylaluminium compounds can exist as either monomers or dimers, depending on the steric bulk of the groups attached. For example, trimethylaluminium exists as a dimer, but trimesitylaluminium adopts a monomeric structure.[6]
Biochemical dimers
Pyrimidine dimers
Pyrimidine dimers (also known as thymine dimers) are formed by a photochemical reaction from pyrimidine DNA bases when exposed to ultraviolet light.[6] This cross-linking causes DNA mutations, which can be carcinogenic, causing skin cancers.[6] When pyrimidine dimers are present, they can block polymerases, decreasing DNA functionality until it is repaired.[6]
Protein dimers
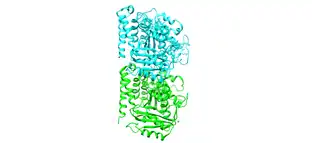
Protein dimers arise from the interaction between two proteins which can interact further to form larger and more complex oligomers.[7] For example, tubulin is formed by the dimerization of α-tubulin and β-tubulin and this dimer can then polymerize further to make microtubules.[8] For symmetric proteins, the larger protein complex can be broken down into smaller identical protein subunits, which then dimerize to decrease the genetic code required to make the functional protein.[7]
G protein-coupled receptors
As the largest and most diverse family of receptors within the human genome, G protein-coupled receptors (GPCR) have been studied extensively, with recent studies supporting their ability to form dimers.[9] GPCR dimers include both homodimers and heterodimers formed from related members of the GPCR family.[10] While not all, some GPCRs require dimerization to function, such as GABAB-receptor, emphasizing the importance of dimers in biological systems.[11]
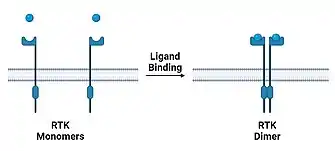
Receptor tyrosine kinase
Much like for G protein-coupled receptors, dimerization is essential for receptor tyrosine kinases (RTK) to perform their function in signal transduction, affecting many different cellular processes.[12] RTKs typically exist as monomers, but undergo a conformational change upon ligand binding, allowing them to dimerize with nearby RTKs.[13][14] The dimerization activates the cytoplasmic kinase domains that are responsible for further signal transduction.[12]
See also
References
- "IUPAC "Gold Book" definition". doi:10.1351/goldbook.D01744. S2CID 242984652. Retrieved 2009-04-30.
{{cite journal}}: Cite journal requires|journal=(help)
- ↑ "Dimerization".
- ↑ Adar, Ram M.; Markovich, Tomer; Andelman, David (2017-05-17). "Bjerrum pairs in ionic solutions: A Poisson-Boltzmann approach". The Journal of Chemical Physics. 146 (19): 194904. arXiv:1702.04853. Bibcode:2017JChPh.146s4904A. doi:10.1063/1.4982885. ISSN 0021-9606. PMID 28527430. S2CID 12227786.
- ↑ Karle, J.; Brockway, L. O. (1944). "An Electron Diffraction Investigation of the Monomers and Dimers of Formic, Acetic and Trifluoroacetic Acids and the Dimer of Deuterium Acetate 1". Journal of the American Chemical Society. 66 (4): 574–584. doi:10.1021/ja01232a022. ISSN 0002-7863.
- ↑ Birks, J B (1975-08-01). "Excimers". Reports on Progress in Physics. 38 (8): 903–974. doi:10.1088/0034-4885/38/8/001. ISSN 0034-4885. S2CID 240065177.
- ↑ Shriver, Duward (2014). Inorganic Chemistry (6th ed.). W.H. Freeman and Company. pp. 306–307. ISBN 9781429299060.
- 1 2 3 4 Shriver, Duward (2014). Inorganic Chemistry (6th ed.). W.H. Freeman and Company. pp. 377–378. ISBN 9781429299060.
- 1 2 Marianayagam, Neelan J.; Sunde, Margaret; Matthews, Jacqueline M. (2004). "The power of two: protein dimerization in biology". Trends in Biochemical Sciences. 29 (11): 618–625. doi:10.1016/j.tibs.2004.09.006. ISSN 0968-0004. PMID 15501681.
- ↑ Cooper, Geoffrey M. (2000). "Microtubules". The Cell: A Molecular Approach. 2nd Edition.
- ↑ Faron-Górecka, Agata; Szlachta, Marta; Kolasa, Magdalena; Solich, Joanna; Górecki, Andrzej; Kuśmider, Maciej; Żurawek, Dariusz; Dziedzicka-Wasylewska, Marta (2019-01-01), Shukla, Arun K. (ed.), "Chapter 10 - Understanding GPCR dimerization", Methods in Cell Biology, G Protein-Coupled Receptors, Part B, Academic Press, 149: 155–178, doi:10.1016/bs.mcb.2018.08.005, ISBN 9780128151075, PMID 30616817, S2CID 58577416, retrieved 2022-10-27
- ↑ Rios, C. D.; Jordan, B. A.; Gomes, I.; Devi, L. A. (2001-11-01). "G-protein-coupled receptor dimerization: modulation of receptor function". Pharmacology & Therapeutics. 92 (2): 71–87. doi:10.1016/S0163-7258(01)00160-7. ISSN 0163-7258. PMID 11916530.
- ↑ Lohse, Martin J (2010-02-01). "Dimerization in GPCR mobility and signaling". Current Opinion in Pharmacology. GPCR. 10 (1): 53–58. doi:10.1016/j.coph.2009.10.007. ISSN 1471-4892. PMID 19910252.
- 1 2 Hubbard, Stevan R (1999-04-01). "Structural analysis of receptor tyrosine kinases". Progress in Biophysics and Molecular Biology. 71 (3): 343–358. doi:10.1016/S0079-6107(98)00047-9. ISSN 0079-6107. PMID 10354703.
- ↑ Lemmon, Mark A.; Schlessinger, Joseph (2010-06-25). "Cell Signaling by Receptor Tyrosine Kinases". Cell. 141 (7): 1117–1134. doi:10.1016/j.cell.2010.06.011. ISSN 0092-8674. PMC 2914105. PMID 20602996.
- ↑ Lemmon, Mark A.; Schlessinger, Joseph; Ferguson, Kathryn M. (2014-04-01). "The EGFR Family: Not So Prototypical Receptor Tyrosine Kinases". Cold Spring Harbor Perspectives in Biology. 6 (4): a020768. doi:10.1101/cshperspect.a020768. ISSN 1943-0264. PMC 3970421. PMID 24691965.